Attorney Docket: 01155-0064-00PCT METHODS OF EDITING AN HLA-A GENE IN VITRO CROSS-REFERENCE TO RELATED APPLICATIONS [001] This application claims the benefit under 35 USC 119(e) of US Provisional Application No.63/579,062, filed August 28, 2023, the content of which is herein incorporated by reference in its entirety. REFERENCE TO ELECTRONIC SEQUENCE LISTING [002] This application contains a sequence listing, which has been submitted electronically in XML file format and is hereby incorporated by reference in its entirety. Said XML file, created on August 13, 2024, is named “01155-0064-00PCT.xml” and is 308,340 bytes in size. INTRODUCTION AND SUMMARY [003] The ability to introduce multiple genetic edits into a cell in vitro is of interest for gene editing and clinical therapeutic applications. For example, adoptive cell therapy approaches using genetically modified cells have become an attractive modality to treat a variety of conditions and diseases, including cancers, to reconstitute cell lineages and immune system defense. However, the clinical application of cell product therapies has been challenging in part due to the complex genetic engineering requirements. The ability to engineer multiple attributes into a single cell depends on the ability to efficiently perform edits in multiple targeted genes, including knockouts and in locus insertions, while avoiding unintended off-target edits. [004] Also, the ability to downregulate MHC class I is of interest for many in vivo and ex vivo utilities, e.g., for creating a cell population in vitro for transplantation. In particular, the transfer of allogeneic cells into a subject is of great interest to the field of cell therapy. The use of allogeneic cells has been limited due to the problem of rejection by the recipient subject’s immune cells, which recognize the transplanted cells as foreign and mount an attack. The ability to downregulate MHC class I can be achieved through reduced or eliminated surface expression of HLA-A. But because of sequence similarities between HLA-A and other HLA alleles, editing of HLA-A may result in off-target edits to such alleles, e.g., HLA-H. Such off- target edits, especially when made as double-strand breaks, may result in chromosome deletion. [005] Provided herein are methods of editing HLA-A in a cell or population of cells in vitro, which are based on the surprising finding that editing HLA-A within certain timeframes, especially relative to activation of the cell or population of cells, reduces the unwanted effects
Attorney Docket: 01155-0064-00PCT (e.g., chromosomal deletion) that results from off-target edits to HLA alleles that share sequence similarities, e.g., HLA-H. The HLA-H may be, for example, HLA-H1, also known as HLA-H*01. BRIEF DESCRIPTION OF THE DRAWINGS [006] Fig. 1 shows the percentage of CD8+ cells positive for the indicated markers in Donor “1” or Donor “2” as measured by flow cytometry. DETAILED DESCRIPTION [007] The present disclosure provides in vitro methods of editing an HLA-A gene in a cell or in a population of cells (e.g., immune cells such as T cells or stem cells such as iPSCs). The methods provide, for example, the ability to edit an HLA-A gene without significant cellular side effects, such as chromosome deletion. The methods also provide, for example, the ability to edit an HLA-A gene without significant loss of viability of the cell or cells. In some embodiments, the methods are manufacturing methods to prepare cells in vitro for subsequent therapeutic administration to a subject. [008] In some embodiments, provided herein is an in vitro method of editing an HLA-A gene in a cell or in a population of cells, comprising the steps of: (a) activating a cell or a population of cells; and (b) contacting the cell or the population of cells at about the same time as, or within about 24 hours before or after, the activating with a first guide RNA (gRNA) that targets an HLA-A gene and a genome editing tool. In some embodiments, the method further comprises the step of: (c) contacting the cell or the population of cells with at least one additional gRNA. In some embodiments, provided herein is an in vitro method of editing an HLA-A gene in a cell or in a population of cells, comprising the steps of: (a) activating the cell or the population of cells; (b) contacting the cell or the population of cells at about the same time as, or within about 24 hours before or after, the activating with a first guide RNA (gRNA) that targets an HLA-A gene and a genome editing tool; and (c) contacting the cell or the population of cells about 3 days after the activating with at least one additional gRNA and a genome editing tool. In some embodiments, the at least one additional gRNA targets a CIITA, TRAC, TRBC, and/or B2M gene. In some embodiments, the at least one additional gRNA targets a CIITA gene. [009] In some embodiments, the cell or population of cells has already been activated and the contacting the cell or population of cells occurs within about 24 hours before or after the activating. In some embodiments, step (c) is performed after the activating and after step (b) described above. In some embodiments, step (c) is performed 3 days after the activating.
Attorney Docket: 01155-0064-00PCT [0010] In some embodiments, the contacting with the first gRNA or the at least one additional gRNA comprises contacting the cell or the population of cells with a lipid nanoparticle (LNP) composition comprising the first gRNA or the at least one additional gRNA. [0011] In some embodiments, provided herein are cell populations comprising edited cells made by or obtainable by the methods disclosed herein. In some embodiments, provided herein are use of the cell populations in a method of therapy or a pharmaceutical composition. In some embodiments, provided herein are methods of creating a cell bank, comprising genetically modifying a cell, using any one of the methods disclosed herein to obtain a population of genetically modified cells, and transferring the genetically modified cells into a cell bank. [0012] In some embodiments, provided herein are cell populations of edited cells, wherein cells in the cell population are homozygous or heterozygous for HLA-H1, wherein no more than 4%, no more than 5%, no more than 6%, no more than 7%, no more than 8%, or no more than 9% of the cells in the cell population are positive for HLA-A surface expression, and wherein (a) a rate of Chr6p21.3 deletion in the cell population is no more than 2 per 200 metaphase spreads, or (b) a copy number variation (CNV) of the short arm of human chromosome 6 in the cell population is no less than 1.90, and/or (c) a rate of Chr6p21.3 deletion and/or a CNV of the short arm of human chromosome 6 in the cell population is no more than a background rate of Chr6p21.3 deletion and/or a background CNV of the short arm of human chromosome 6, respectively, in a same but unedited cell population. [0013] Provided herein are the following numbered embodiments: [0014] Embodiment 1 is an in vitro method of editing an HLA-A gene in a cell or in a population of cells, comprising the steps of: (a) activating the cell or the population of cells; and (b) contacting the cell or the population of cells at about the same time as, or within about 24 hours before or after, the activating with a first guide RNA (gRNA) that targets an HLA-A gene and a genome editing tool. [0015] Embodiment 2 is the method of embodiment 1, wherein the cell or population of cells has already been activated and wherein contacting the cell or population of cells occurs within about 24 hours before or after the activating. [0016] Embodiment 3 is the method of embodiment 1 or 2, further comprising the step of: (c) contacting the cell or the population of cells with at least one additional gRNA. [0017] Embodiment 4 is the method of embodiment 3, wherein the at least one additional gRNA targets a CIITA, TRAC, TRBC, and/or B2M gene.
Attorney Docket: 01155-0064-00PCT [0018] Embodiment 5 is the method of embodiment 3, wherein the at least one additional gRNA targets a CIITA gene. [0019] Embodiment 6 is the method of any one of embodiments 3-5, wherein step (c) is performed after the activating and after step (b). [0020] Embodiment 7 is the method of any one of embodiments 3-6, wherein step (c) is performed 1, 2, or 3 days after the activating. [0021] Embodiment 8 is the method of any one of embodiments 3-6, wherein step (c) is performed 3 days after the activating. [0022] Embodiment 9 is an in vitro method of editing an HLA-A gene in a cell or in a population of cells, comprising the steps of: (a) activating the cell or the population of cells; (b) contacting the cell or the population of cells at about the same time as, or within about 24 hours before or after, the activating with a first guide RNA (gRNA) that targets an HLA-A gene and a genome editing tool; and (c) contacting the cell or the population of cells about 3 days after the activating with at least one additional gRNA and a genome editing tool. [0023] Embodiment 10 is the method of embodiment 9, wherein the at least one additional gRNA targets a CIITA, TRAC, TRBC, and/or B2M gene. [0024] Embodiment 11 is the method of embodiment 9, wherein the at least one additional gRNA targets a CIITA gene. [0025] Embodiment 12 is the method of any one of embodiments 1-11, wherein the contacting with the first gRNA or the at least one additional gRNA comprises contacting the cell or the population of cells with a lipid nanoparticle (LNP) composition comprising the first gRNA or the at least one additional gRNA. [0026] Embodiment 13 is the method of embodiment 12, wherein the LNP further comprises a nucleic acid encoding the genome editing tool. [0027] Embodiment 14 is the method of any one of embodiments 1-13, wherein the genome editing tool is an RNA-guided DNA binding agent or is a nucleic acid genome editing tool encoding an RNA-guided DNA binding agent. [0028] Embodiment 15 is the method of embodiment 14, wherein the nucleic acid genome editing tool is an mRNA encoding the RNA-guided DNA binding agent. [0029] Embodiment 16 is the method of embodiment 15, wherein the mRNA encoding the RNA-guided DNA binding agent comprises an open reading frame (ORF) comprising any one of SEQ ID NOs: 1-5, 29, and 30 or a sequence having at least 90% identity to any one of SEQ ID NOs: 1-5, 29, and 30.
Attorney Docket: 01155-0064-00PCT [0030] Embodiment 17 is the method of any one of embodiments 14-16, wherein the RNA- guided DNA binding agent is a cleavase. [0031] Embodiment 18 is the method of any one of embodiments 14-17, wherein the RNA- guided DNA binding agent is a Cas9. [0032] Embodiment 19 is the method of embodiment 18, wherein the RNA-guided DNA binding agent is an S. pyogenes Cas9. [0033] Embodiment 20 is the method of any one of embodiments 1-19, wherein the cell is a human cell. [0034] Embodiment 21 is the method of any one of embodiments 1-20, wherein the cell is an immune cell. [0035] Embodiment 22 is the method of embodiment 21, wherein the immune cell is a lymphocyte, a T cell, a B cell, a natural killer cell, a natural killer T cell, a monocyte, a macrophage, a mast cell, a dendritic cell, a granulocyte (e.g., neutrophil, eosinophil, and basophil), a primary immune cell, a CD3+ cell, a CD4+ cell, a CD8+ T cell, or a regulatory T cell (Treg). [0036] Embodiment 23 is the method of embodiment 21 or 22, wherein the immune cell is a T cell. [0037] Embodiment 24 is the method of any one of embodiments 21-23, wherein the immune cell is a natural killer cell. [0038] Embodiment 25 is the method of any one of embodiments 1-20, wherein the cell is a stem cell. [0039] Embodiment 26 is the method of embodiment 25, wherein the stem cell is a mesenchymal stem cell, a hematopoietic stem cell (HSC), a neural stem cells (NSC), a limbal stem cell (LSC), an induced pluripotent stem cell (iPSC), an ocular stem cell, a pluripotent stem cell (PSC), or an embryonic stem cell (ESC). [0040] Embodiment 27 is the method of embodiment 25 or 26, wherein the stem cell is an iPSC. [0041] Embodiment 28 is the method of any one of embodiments 1-27, wherein the cell or the population of cells is cultured, expanded, or proliferated ex vivo either before or after gene editing. [0042] Embodiment 29 is the method of any one of embodiments 1-28, wherein the first gRNA or the at least one additional gRNA comprises a single guide RNA (sgRNA). [0043] Embodiment 30 is the method of any one of embodiments 1-29, wherein the first gRNA or the at least one additional gRNA comprises a dual guide RNA (dgRNA).
Attorney Docket: 01155-0064-00PCT [0044] Embodiment 31 is the method of any one of embodiments 1-30, wherein the first gRNA comprises the sequence of SEQ ID NO: 727, a sequence having 90% or 95% identity to SEQ ID NO: 727, or a sequence having at least 17, 18, 19, or 20 contiguous nucleotides of SEQ ID NO: 727. [0045] Embodiment 32 is the method of any one of embodiments 1-31, wherein the first gRNA comprises the sequence of SEQ ID NO: 716. [0046] Embodiment 33 is the method of any one of embodiments 1-32, wherein the at least one additional gRNA comprises the sequence of SEQ ID NO: 728, a sequence having 90% or 95% identity to SEQ ID NO: 728, or a sequence having at least 17, 18, 19, or 20 contiguous nucleotides of SEQ ID NO: 728. [0047] Embodiment 34 is the method of any one of embodiments 1-33, wherein the at least one additional gRNA comprises the sequence of SEQ ID NO: 724. [0048] Embodiment 35 is the method of any one of embodiments 1-34, wherein the cell or population of cells is contacted with a cleavase and no more than two guide RNAs simultaneously. [0049] Embodiment 36 is the method of any one of embodiments 1-35, further comprising contacting the cell or the population of cells with a DNA-dependent protein kinase inhibitor (DNA-PKi). [0050] Embodiment 37 is the method of embodiment 36, wherein the DNA-PKi is selected from Compound 1 and Compound 4. [0051] Embodiment 38 is the method of any one of embodiments 1-37, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids. [0052] Embodiment 39 is the method of any one of embodiments 1-38, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein the one or more donor nucleic acids comprise a vector. [0053] Embodiment 40 is the method of any one of embodiments 1-39, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein the one or more donor nucleic acids comprise a viral vector. [0054] Embodiment 41 is the method of any one of embodiments 1-40, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein the one or more donor nucleic acids comprise a lentiviral vector or optionally a retroviral vector.
Attorney Docket: 01155-0064-00PCT [0055] Embodiment 42 is the method of any one of embodiments 1-41, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein the one or more donor nucleic acids comprise an AAV vector. [0056] Embodiment 43 is the method of any one of embodiments 1-42, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids is provided in a LNP composition, or wherein the method further comprises contacting the cell or the population of cells with a LNP composition comprising one or more donor nucleic acids. [0057] Embodiment 44 is the method of any one of embodiments 1-43, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids is integrated into the cell or population of cells by homologous recombination. [0058] Embodiment 45 is the method of any one of embodiments 1-44, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids comprise flanking nucleic acid regions homologous to all or part of the target sequence. [0059] Embodiment 46 is the method of any one of embodiments 1-45, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids is integrated into the cell or population of cells by blunt end insertion. [0060] Embodiment 47 is the method of any one of embodiments 1-46, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids is integrated into the cell or population of cells by non-homologous end joining. [0061] Embodiment 48 is the method of any one of embodiments 1-47, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein the one or more donor nucleic acids is inserted into a safe harbor locus in the cell or population of cells. [0062] Embodiment 49 is the method of any one of embodiments 1-48, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids comprises regions having homology with corresponding regions of a T cell receptor sequence in the cell or population of cells, wherein the homology allows integration of the at least one of the one or more donor nucleic acids into the T cell receptor sequence.
Attorney Docket: 01155-0064-00PCT [0063] Embodiment 50 is the method of any one of embodiments 1-49, wherein the method further comprises contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids comprises regions having homology with corresponding regions of a TRAC locus, a B2M locus, an AAVS1 locus, and/or CIITA locus, or optionally a TRBC locus in the cell or population of cells, wherein the homology allows integration of the at least one of the one or more donor nucleic acids into the locus or loci. [0064] Embodiment 51 is the method of any one of embodiments 1-50, further comprising contacting the cell or the population of cells with a gRNA targeting TRAC. [0065] Embodiment 52 is the method of any one of embodiments 1-51, further comprising contacting the cell or the population of cells with a gRNA targeting TRBC. [0066] Embodiment 53 is the method of any one of embodiments 1-52, further comprising contacting the cell or the population of cells with a gRNA targeting B2M. [0067] Embodiment 54 is the method of any one of embodiments 1-53, wherein the cell or the population of cells has reduced or eliminated surface expression of HLA-A and HLA-B, and is homozygous for HLA-C. [0068] Embodiment 55 is the method of any one of embodiments 1-54, wherein the cell or the population of cells is homozygous for HLA-B and homozygous for HLA-C. [0069] Embodiment 56 is the method of any one of embodiments 1-55, wherein the cell or the population of cells has reduced or eliminated surface expression of HLA-A, and is homozygous for HLA-B and homozygous for HLA-C. [0070] Embodiment 57 is the method of any one of embodiments 1-56, wherein the cell or the population of cells is HLA-H1 positive and/or is homozygous or heterozygous for HLA- H1. [0071] Embodiment 58 is the method of any one of embodiments 1-57, wherein the method further comprises contacting the cell or the population of cells with at least two LNP compositions, wherein at least one of the LNP compositions comprises a gRNA targeting a gene that reduces or eliminates surface expression of MHC class II. [0072] Embodiment 59 is the method of any one of embodiments 1-58, wherein the method further comprises contacting the cell or the population of cells with at least two LNP compositions, wherein at least one of the LNP compositions comprises a gRNA targeting TRAC, and at least one of the LNP compositions comprises a gRNA targeting TRBC. [0073] Embodiment 60 is the method of embodiment 59, wherein at least one of the LNP compositions comprises a gRNA targeting B2M.
Attorney Docket: 01155-0064-00PCT [0074] Embodiment 61 is the method of any one of embodiments 1-60, wherein the method does not include a selection step. [0075] Embodiment 62 is the method of any one of embodiments 1-60, wherein the method comprises a selection step, wherein the selection step is a physical sorting step or a biochemical selecting step. [0076] Embodiment 63 is a cell population made by or obtainable by the method of any one of embodiments 1-62. [0077] Embodiment 64 is the cell population of embodiment 63, wherein at least 70% of the cells are viable 24 hours after contacting the population of cells with an LNP composition. [0078] Embodiment 65 is the cell population of embodiment 63 or 64, wherein the population comprises T cells and wherein at least 95% of the cells in the population comprises a genome edit of an endogenous T cell receptor (TCR) sequence. [0079] Embodiment 66 is the cell population of any one of embodiments 63-65, wherein the population comprises T cells and wherein at least 30%, 40%, optionally 50%, 55%, 60%, 65% of the cells of the population of cells has a memory phenotype (CD45RA+/CD27+). [0080] Embodiment 67 is the cell population of any one of embodiments 63-66, wherein the population comprises T cells and is responsive to repeat stimulation after editing. [0081] Embodiment 68 is the cell population of any one of embodiments 63-67, wherein the population comprises a genome edit comprising insertion of a heterologous sequence coding for a targeting ligand or an alternative antigen binding moiety in 70%, 75%, 80%, or 85% of the cells of the population. [0082] Embodiment 69 is the cell population of any one of embodiments 63-68, wherein (a) a rate of Chr6p21.3 deletion in the cell population is no more than 2 per 200 metaphase spreads, or (b) a copy number variation (CNV) of the short arm of human chromosome 6 in the cell population is no less than 1.90, and/or (c) a rate of Chr6p21.3 deletion and/or a CNV of the short arm of human chromosome 6 in the population is no more than a background rate of Chr6p21.3 deletion and/or a background CNV of the short arm of human chromosome 6, respectively, in a same cell population on which the method has not been performed. [0083] Embodiment 70 is a cell population of edited cells, wherein cells in the cell population are homozygous or heterozygous for HLA-H1, wherein no more than 4%, no more than 5%, no more than 6%, no more than 7%, no more than 8%, or no more than 9% of the cells in the cell population are positive for HLA-A surface expression, and wherein (a) a rate of Chr6p21.3 deletion in the cell population is no more than 2 per 200 metaphase spreads, or (b) a copy number variation (CNV) of the short arm of human chromosome 6 in the cell
Attorney Docket: 01155-0064-00PCT population is no less than 1.90, and/or (c) a rate of Chr6p21.3 deletion and/or a CNV of the short arm of human chromosome 6 in the cell population is no more than a background rate of Chr6p21.3 deletion and/or a background CNV of the short arm of human chromosome 6, respectively, in a same but unedited cell population. [0084] Embodiment 71 is the cell population of embodiment 70, wherein the HLA-A surface expression is measured by flow cytometry. [0085] Embodiment 72 is the cell population of any one of embodiments 69-71, wherein the rate of Chr6p21.3 deletion is measured by a karyotype analysis. [0086] Embodiment 73 is the cell population of any one of embodiments 69-72, wherein the CNV is measured using a NEDD9 gene as a marker for the distal portion of the short arm of human chromosome 6 and using a MRPL18 gene as a reference control on the long arm of human chromosome 6. [0087] Embodiment 74 is the cell population of any one of embodiments 69-73, wherein the CNV is measured by droplet digital PCR. [0088] Embodiment 75 is the cell population of any one of embodiments 70-74, wherein the edited cells in the cell population comprise a genetic modification in a HLA-A gene, wherein the genetic modification comprises at least one nucleotide within the genomic coordinates chr6:29942864-29942884. [0089] Embodiment 76 is the cell population of embodiment 75, wherein the genetic modification in the HLA-A gene is created by a cleavase. [0090] Embodiment 77 is the cell population of embodiment 76, wherein the cleavase is an S. pyogenes Cas9 cleavase. [0091] Embodiment 78 is the cell population of any one of embodiments 70-77, wherein the edited cells in the cell population comprise a genetic modification in a CIITA, TRAC, TRBC, and/or B2M gene. [0092] Embodiment 79 is the cell population of any one of embodiments 70-78, wherein the edited cells in the cell population comprise a genetic modification in the CIITA gene. [0093] Embodiment 80 is the cell population of any one of embodiments 70-79, wherein the edited cells in the cell population comprise a genetic modification in the TRAC gene. [0094] Embodiment 81 is the cell population of any one of embodiments 70-80, wherein the edited cells in the cell population comprise insertion of a heterologous sequence coding for a targeting ligand or an alternative antigen. [0095] Embodiment 82 is the cell population of any one of embodiments 70-81, wherein the edited cells comprise lymphocytes, T cells, B cells, natural killer cells, natural killer T cells,
Attorney Docket: 01155-0064-00PCT monocytes, macrophages, mast cells, dendritic cells, granulocytes, primary immune cells, CD3+ cells, CD4+ cells, CD8+ cells, and/or regulatory T cells. [0096] Embodiment 83 is the cell population of any one of embodiments 70-82, wherein the edited cells comprise T cells. [0097] Embodiment 84 is the cell population of any one of embodiments 70-83, wherein the edited cells comprise CD4+ T cells. [0098] Embodiment 85 is the cell population of any one of embodiments 70-83, wherein the edited cells comprise CD8+ T cells. [0099] Embodiment 86 is the cell population of any one of embodiments 70-82, wherein the edited cells comprise natural killer cells. [00100] Embodiment 87 is the cell population of any one of embodiments 70-86, wherein the edited cells comprise activated cells. [00101] Embodiment 88 is the cell population of any one of embodiments 70-87, wherein the edited cells are homozygous for HLA-B and/or homozygous for HLA-C. [00102] Embodiment 89 is a pharmaceutical composition comprising the cell population of any one of embodiments 63-88. [00103] Embodiment 90 is use of the cell population of any one of embodiments 63-88 or the pharmaceutical composition of embodiment 89, in a method of therapy. [00104] Embodiment 91 is the use of the cell population or pharmaceutical composition of embodiment 90, wherein the method of therapy is for treatment of cancer or autoimmune therapy. [00105] Embodiment 92 is the use of the cell population or pharmaceutical composition of embodiment 90 or 91, wherein the method of therapy is for adoptive cell transfer therapy. [00106] Embodiment 93 is a method of treating cancer, a method of treating an autoimmune disorder, or a method of adoptive cell transfer therapy, comprising administering to a subject in need thereof a therapeutically effective amount of the cell population of any one of embodiments 63-88 or the pharmaceutical composition of embodiment 89. [00107] Embodiment 94 is a method of creating a cell bank, comprising genetically modifying a cell, using the method according to any of embodiments 1-62 to obtain a population of genetically modified cells, and transferring the genetically modified cells into a cell bank.
Attorney Docket: 01155-0064-00PCT I. Definitions [00108] Unless stated otherwise, the following terms and phrases as used herein are intended to have the following meanings: [00109] “Polynucleotide” and “nucleic acid” are used herein to refer to a multimeric compound comprising nucleosides or nucleoside analogs which have nitrogenous heterocyclic bases or base analogs linked together along a backbone, including conventional RNA, DNA, mixed RNA-DNA, and polymers that are analogs thereof. A nucleic acid “backbone” can be made up of a variety of linkages, including one or more of sugar-phosphodiester linkages, peptide-nucleic acid bonds (“peptide nucleic acids” or PNA; PCT No. WO 95/32305), phosphorothioate linkages, methylphosphonate linkages, or combinations thereof. Sugar moieties of a nucleic acid can be ribose, deoxyribose, or similar compounds with substitutions, e.g., 2’ methoxy or 2’ halide substitutions. Nitrogenous bases can be conventional bases (A, G, C, T, U), analogs thereof (e.g., modified uridines such as 5-methoxyuridine, pseudouridine, or N1-methylpseudouridine, or others); inosine; derivatives of purines or pyrimidines (e.g., N
4- methyl deoxyguanosine, deaza- or aza-purines, deaza- or aza-pyrimidines, pyrimidine bases with substituent groups at the 5 or 6 position (e.g., 5-methylcytosine), purine bases with a substituent at the 2, 6, or 8 positions, 2-amino-6-methylaminopurine, O
6-methylguanine, 4- thio-pyrimidines, 4-amino-pyrimidines, 4-dimethylhydrazine-pyrimidines, and O
4-alkyl- pyrimidines; US Pat. No. 5,378,825 and PCT No. WO 93/13121). For general discussion see The Biochemistry of the Nucleic Acids 5-36, Adams et al., ed., 11
th ed., 1992). Nucleic acids can include one or more “abasic” residues where the backbone includes no nitrogenous base for position(s) of the polymer (US Pat. No. 5,585,481). A nucleic acid can comprise only conventional RNA or DNA sugars, bases and linkages, or can include both conventional components and substitutions (e.g., conventional bases with 2’ methoxy linkages, or polymers containing both conventional bases and one or more base analogs). Nucleic acid includes “locked nucleic acid” (LNA), an analogue containing one or more LNA nucleotide monomers with a bicyclic furanose unit locked in an RNA mimicking sugar conformation, which enhance hybridization affinity toward complementary RNA and DNA sequences (Vester and Wengel, 2004, Biochemistry 43(42):13233-41). RNA and DNA have different sugar moieties and can differ by the presence of uracil or analogs thereof in RNA and thymine or analogs thereof in DNA. [00110] “Guide RNA”, “gRNA”, and simply “guide” are used herein interchangeably to refer to the guide that directs an RNA-guided DNA binding agent to a target DNA and can be
Attorney Docket: 01155-0064-00PCT either a crRNA (also known as CRISPR RNA), or the combination of a crRNA and a trRNA (also known as tracrRNA). The crRNA and trRNA may be associated as a single RNA molecule (single guide RNA, sgRNA) or in two separate RNA molecules (dual guide RNA, dgRNA). “Guide RNA” or “gRNA” refers to each type. The trRNA may be a naturally occurring sequence, or a trRNA sequence with modifications or variations compared to naturally-occurring sequences. [00111] As used herein, a “guide sequence” refers to a sequence within a guide RNA that is complementary to a target sequence and functions to direct a guide RNA to a target sequence for binding or modification (e.g., cleavage) by an RNA-guided DNA binding agent. A “guide sequence” may also be referred to as a “targeting sequence,” or a “spacer sequence.” A guide sequence can be 20 base pairs in length, e.g., in the case of Streptococcus pyogenes (i.e., Spy Cas9) and related Cas9 homologs/orthologs. Shorter or longer sequences can also be used as guides, e.g., 15-, 16-, 17-, 18-, 19-, 21-, 22-, 23-, 24-, or 25-nucleotides in length. In some embodiments, the target sequence is in a gene or on a chromosome, for example, and is complementary to the guide sequence. In some embodiments, the degree of complementarity or identity between a guide sequence and its corresponding target sequence may be about 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99%, or 100%. In some embodiments, the guide sequence and the target region may be 100% complementary or identical. In other embodiments, the guide sequence and the target region may contain at least one mismatch. For example, the guide sequence and the target sequence may contain 1, 2, 3, or 4 mismatches, where the total length of the target sequence is at least 17, 18, 19, 20 or more base pairs. In some embodiments, the guide sequence and the target region may contain 1-4 mismatches where the guide sequence comprises at least 17, 18, 19, 20 or more nucleotides. In some embodiments, the guide sequence and the target region may contain 1, 2, 3, or 4 mismatches where the guide sequence comprises 20 nucleotides. [00112] Target sequences for RNA-guided DNA binding agents include both the positive and negative strands of genomic DNA (i.e., the sequence given and the sequence’s reverse compliment), as a nucleic acid substrate for an RNA-guided DNA binding agent is a double stranded nucleic acid. Accordingly, where a guide sequence is said to be “complementary to a target sequence”, it is to be understood that the guide sequence may direct a guide RNA to bind to the reverse complement of a target sequence. Thus, in some embodiments, where the guide sequence binds the reverse complement of a target sequence, the guide sequence is identical to certain nucleotides of the target sequence (e.g., the target sequence not including the PAM) except for the substitution of U for T in the guide sequence.
Attorney Docket: 01155-0064-00PCT [00113] As used herein, an “RNA-guided DNA binding agent” means a polypeptide or complex of polypeptides having RNA and DNA binding activity, or a DNA-binding subunit of such a complex, wherein the DNA binding activity is sequence-specific and depends on the sequence of the RNA. Exemplary RNA-guided DNA binding agents include Cas cleavases/nickases and inactivated forms thereof (“dCas DNA binding agents”). “Cas nuclease”, also called “Cas protein” as used herein, encompasses Cas cleavases, Cas nickases, and dCas DNA binding agents. Cas cleavases/nickases and dCas DNA binding agents include a Csm or Cmr complex of a type III CRISPR system, the Cas10, Csm1, or Cmr2 subunit thereof, a Cascade complex of a type I CRISPR system, the Cas3 subunit thereof, and Class 2 Cas nucleases. As used herein, a “Class 2 Cas nuclease” is a single-chain polypeptide with RNA-guided DNA binding activity. Class 2 Cas nucleases include Class 2 Cas cleavases/nickases (e.g., H840A, D10A, or N863A variants), which further have RNA-guided DNA cleavases or nickase activity, and Class 2 dCas DNA binding agents, in which cleavase/nickase activity is inactivated. Class 2 Cas nucleases include, for example, Cas9, Cpf1, C2c1, C2c2, C2c3, HF Cas9 (e.g., N497A, R661A, Q695A, Q926A variants), HypaCas9 (e.g., N692A, M694A, Q695A, H698A variants), eSPCas9(1.0) (e.g., K810A, K1003A, R1060A variants), and eSPCas9(1.1) (e.g., K848A, K1003A, R1060A variants) proteins and modifications thereof. Cpf1 protein, Zetsche et al., Cell, 163: 1-13 (2015), is homologous to Cas9, and contains a RuvC-like nuclease domain. Cpf1 sequences of Zetsche are incorporated by reference in their entirety. See, e.g., Zetsche, Tables S1 and S3. See, e.g., Makarova et al., Nat Rev Microbiol, 13(11): 722-36 (2015); Shmakov et al., Molecular Cell, 60:385-397 (2015). [00114] As used herein, “ribonucleoprotein” (RNP) or “RNP complex” refers to a guide RNA together with an RNA-guided DNA binding agent, such as a Cas nuclease, e.g., a Cas cleavase, Cas nickase, or dCas DNA binding agent (e.g., Cas9). In some embodiments, the guide RNA guides the RNA-guided DNA binding agent such as Cas9 to a target sequence, and the guide RNA hybridizes with and the agent binds to the target sequence; in cases where the agent is a cleavase or nickase, binding can be followed by cleaving or nicking. [00115] As used herein, the term “editor” refers to an agent comprising a polypeptide that is capable of making a modification to a base (e.g., A, T, C, G, or U) within a nucleic acid sequence (e.g., DNA or RNA). In some embodiments, the editor is capable of deaminating a base within a nucleic acid. In some embodiments, the editor is capable of deaminating a base within a DNA molecule. In some embodiments, the editor is capable of deaminating a cytosine (C) in DNA. In some embodiments, the editor is a fusion protein comprising an RNA-guided nickase fused to a cytidine deaminase domain. In some embodiments, the editor is a fusion
Attorney Docket: 01155-0064-00PCT protein comprising an RNA-guided nickase fused to an APOBEC3A deaminase (A3A). In some embodiments, the editor comprises a Cas9 nickase fused to an APOBEC3A deaminase (A3A). [00116] As used herein, a first sequence is considered to “comprise a sequence with at least X% identity to” a second sequence if an alignment of the first sequence to the second sequence shows that X% or more of the positions of the second sequence in its entirety are matched by the first sequence. For example, the sequence AAGA comprises a sequence with 100% identity to the sequence AAG because an alignment would give 100% identity in that there are matches to all three positions of the second sequence. The differences between RNA and DNA (generally the exchange of uridine for thymidine or vice versa) and the presence of nucleoside analogs such as modified uridines do not contribute to differences in identity or complementarity among polynucleotides as long as the relevant nucleotides (such as thymidine, uridine, or modified uridine) have the same complement (e.g., adenosine for all of thymidine, uridine, or modified uridine; another example is cytosine and 5-methylcytosine, both of which have guanosine or modified guanosine as a complement). Thus, for example, the sequence 5’-AXG where X is any modified uridine, such as pseudouridine, N1-methyl pseudouridine, or 5-methoxyuridine, is considered 100% identical to AUG in that both are perfectly complementary to the same sequence (5’-CAU). Exemplary alignment algorithms are the Smith-Waterman and Needleman-Wunsch algorithms, which are well-known in the art. One skilled in the art will understand what choice of algorithm and parameter settings are appropriate for a given pair of sequences to be aligned; for sequences of generally similar length and expected identity >50% for amino acids or >75% for nucleotides, the Needleman- Wunsch algorithm with default settings of the Needleman-Wunsch algorithm interface provided by the EBI at the www.ebi.ac.uk web server is generally appropriate. [00117] “mRNA” is used herein to refer to a polynucleotide and comprises an open reading frame that can be translated into a polypeptide (i.e., can serve as a substrate for translation by a ribosome and amino-acylated tRNAs). mRNA can comprise a phosphate-sugar backbone including ribose residues or analogs thereof, e.g., 2’-methoxy ribose residues. In some embodiments, the sugars of an mRNA phosphate-sugar backbone consist essentially of ribose residues, 2’-methoxy ribose residues, or a combination thereof. [00118] As used herein, “indels” refer to insertion/deletion mutations consisting of a number of nucleotides that are either inserted or deleted, e.g., at the site of double-stranded breaks (DSBs) in a target nucleic acid.
Attorney Docket: 01155-0064-00PCT [00119] As used herein, “knockdown” refers to a decrease in expression of a particular gene product (e.g., protein, mRNA, or both). Knockdown of a protein can be measured by detecting total cellular amount of the protein from a sample, such as a tissue, fluid, or cell population of interest. It can also be measured by measuring a surrogate, marker, or activity for the protein. Methods for measuring knockdown of mRNA are known and include sequencing of mRNA isolated from a sample of interest. In some embodiments, “knockdown” may refer to some loss of expression of a particular gene product, for example a decrease in the amount of mRNA transcribed or a decrease in the amount of protein expressed by a population of cells (including in vivo populations such as those found in tissues). [00120] As used herein, “knockout” refers to a loss of expression from a particular gene or of a particular protein in a cell. Knockout can be measured either by detecting total cellular amount of a protein in a cell, a tissue or a population of cells. [00121] As used herein, a “target sequence” refers to a sequence of nucleic acid in a target gene that has complementarity to the guide sequence of the gRNA. The interaction of the target sequence and the guide sequence directs an RNA-guided DNA binding agent to bind, and potentially nick or cleave (depending on the activity of the agent), within the target sequence. [00122] As used herein, “treatment” refers to any administration or application of a therapeutic for disease or disorder in a subject, and includes inhibiting the disease, arresting its development, relieving one or more symptoms of the disease, curing the disease, or preventing one or more symptoms of the disease, including reoccurrence of the symptom. [00123] As used herein, a “cell population comprising edited cells,” or a “population of cells comprising edited cells,” or the like refers to a cell population that comprises edited cells, however not all cells in the population must be edited. A cell population comprising edited cells may also include non-edited cells. The percentage of edited cells within a cell population comprising edited cells may be determined by counting the number of cells within the population that are edited in the population as determined by standard cell counting methods. For example, in some embodiments, a cell population comprising edited cells comprising a single genome edit will have at least 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 99% of the cells in the population with the single edit. In some embodiments, a cell population comprising edited cells comprising at least two genome edits will have at least 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, or 95% of the cells in the population with at least two genome edits. [00124] The term “about” or “approximately” means an acceptable error for a particular value as determined by one of ordinary skill in the art, which depends in part on how the value
Attorney Docket: 01155-0064-00PCT is measured or determined, or a degree of variation that does not substantially affect the properties of the described subject matter, or within the tolerances accepted in the art, e.g., within 10%, 5%, 2%, or 1%. Accordingly, unless indicated to the contrary, the numerical parameters set forth in the following specification and attached claims are approximations that may vary depending upon the desired properties sought to be obtained. At the very least, and not as an attempt to limit the application of the doctrine of equivalents to the scope of the claims, each numerical parameter should at least be construed in light of the number of reported significant digits and by applying ordinary rounding techniques. [00125] Reference will now be made in detail to certain embodiments of the invention, examples of which are illustrated in the accompanying drawings. While the invention is described in conjunction with the illustrated embodiments, it will be understood that they are not intended to limit the invention to those embodiments. On the contrary, the invention is intended to cover all alternatives, modifications, and equivalents, which may be included within the invention as defined by the appended claims and included embodiments. [00126] Before describing the present teachings in detail, it is to be understood that the disclosure is not limited to specific compositions or process steps, as such may vary. It should be noted that, as used in this specification and the appended claims, the singular form “a”, “an” and “the” include plural references unless the context clearly dictates otherwise. Thus, for example, reference to “a conjugate” includes a plurality of conjugates and reference to “a cell” includes a plurality of cells and the like. [00127] Numeric ranges are inclusive of the numbers defining the range. Measured and measurable values are understood to be approximate, taking into account significant digits and the error associated with the measurement. Also, the use of “comprise”, “comprises”, “comprising”, “contain”, “contains”, “containing”, “include”, “includes”, and “including” are not intended to be limiting. It is to be understood that both the foregoing general description and detailed description are exemplary and explanatory only and are not restrictive of the teachings. [00128] Unless specifically noted in the specification, embodiments in the specification that recite “comprising” various components are also contemplated as “consisting of” or “consisting essentially of” the recited components; embodiments in the specification that recite “consisting of” various components are also contemplated as “comprising” or “consisting essentially of” the recited components; and embodiments in the specification that recite “consisting essentially of” various components are also contemplated as “consisting of” or “comprising” the recited components (this interchangeability does not apply to the use of these terms in the claims). The
Attorney Docket: 01155-0064-00PCT term “or” is used in an inclusive sense, i.e., equivalent to “and/or,” unless the context clearly indicates otherwise. [00129] The section headings used herein are for organizational purposes only and are not to be construed as limiting the desired subject matter in any way. In the event that any material incorporated by reference contradicts any term defined in this specification or any other express content of this specification, this specification controls. While the present teachings are described in conjunction with various embodiments, it is not intended that the present teachings be limited to such embodiments. On the contrary, the present teachings encompass various alternatives, modifications, and equivalents, as will be appreciated by those of skill in the art. II. Gene Editing [00130] In some embodiments, provided herein is an in vitro method of editing an HLA-A gene in a cell or in a population of cells, comprising the steps of: (a) activating a cell or a population of cells; and (b) contacting the cell or the population of cells at about the same time as, or within about 24 hours before or after, the activating with a first guide RNA (gRNA) that targets an HLA-A gene and a genome editing tool. In some embodiments, provided herein is an in vitro method of editing an HLA-A gene in a cell or in a population of cells, comprising the steps of: (a) activating the cell or the population of cells; (b) contacting the cell or the population of cells at about the same time as, or within about 24 hours before or after, the activating with a first guide RNA (gRNA) that targets an HLA-A gene and a genome editing tool; and (c) contacting the cell or the population of cells about 3 days after the activating with at least one additional gRNA and a genome editing tool. The cell or the population of cells may be an immune cell (any one of the immune cells or populations of immune cells described herein) or a stem cell (any one of the stem cells or populations of stem cells described herein, e.g., iPSC). The genome editing tool may be any one of the genome editing tools described herein. [00131] In some embodiments, the cell or population of cells has already been activated and the contacting the cell or population of cells occurs within about 24 hours before or after the activating. In some embodiments, the cell or population of cells has already been activated and the contacting the cell or population of cells occurs within about 24 hours before the activating. In some embodiments, the cell or population of cells has already been activated and the contacting the cell or population of cells occurs within about 24 hours after the activating. In some embodiments, the cell or population of cells has already been activated and the contacting the cell or population of cells occurs about 24 hours before the activating. In some
Attorney Docket: 01155-0064-00PCT embodiments, the cell or population of cells has already been activated and the contacting the cell or population of cells occurs about 24 hours after the activating. [00132] In some embodiments, the methods described herein further comprise the step of: (c) contacting the cell or the population of cells with at least one additional gRNA. In some embodiments, the at least one additional gRNA targets a CIITA, TRAC, TRBC, and/or B2M gene. In some embodiments, the at least one additional gRNA targets a CIITA gene. In some embodiments, the at least one additional gRNA targets a TRAC gene. In some embodiments, the at least one additional gRNA targets a TRBC gene. In some embodiments, the at least one additional gRNA targets a B2M gene. In some embodiments, the at least one additional gRNA targets a CIITA gene and a TRAC gene. [00133] In some embodiments, step (c) described above is performed after the activating and after step (b) described above. In some embodiments, step (c) is performed 1, 2, or 3 days after the activating. In some embodiments, step (c) is performed 1 days after the activating. In some embodiments, step (c) is performed 2 days after the activating. In some embodiments, step (c) is performed 3 days after the activating. [00134] In some embodiments, the contacting with the first gRNA or the at least one additional gRNA comprises contacting the cell or the population of cells with a lipid nanoparticle (LNP) composition comprising the first gRNA or the at least one additional gRNA. In some embodiments, the contacting with the first gRNA comprises contacting the cell or the population of cells with an LNP composition comprising the first gRNA. In some embodiments, the contacting with the at least one additional gRNA comprises contacting the cell or the population of cells with an LNP composition comprising the at least one additional gRNA. [00135] In some embodiments, the cell or population of cells is contacted with a cleavase and no more than two guide RNAs simultaneously. [00136] In some embodiments, the methods disclosed herein further comprise contacting the cell or the population of cells with one or more donor nucleic acids, wherein at least one of the one or more donor nucleic acids is provided in a LNP composition. In some embodiments, the methods disclosed herein further comprise contacting the cell or the population of cells with a LNP composition comprising one or more donor nucleic acids. In some embodiments, the methods disclosed herein further comprise contacting the cell or the population of cells with at least two LNP compositions, wherein at least one of the LNP compositions comprises a gRNA targeting a gene that reduces or eliminates surface expression of MHC class II. In some embodiments, the methods disclosed herein further comprise contacting the cell or the
Attorney Docket: 01155-0064-00PCT population of cells with at least two LNP compositions, wherein at least one of the LNP compositions comprises a gRNA targeting TRAC, and at least one of the LNP compositions comprises a gRNA targeting TRBC. In some embodiments, at least one of the LNP compositions comprises a gRNA targeting B2M. [00137] β2M or B2M are used interchangeably herein and with reference to nucleic acid sequence or protein sequence of β-2 microglobulin; the human gene has accession number NC_000015 (range 44711492..44718877), reference GRCh38.p13. The B2M protein is associated with MHC class I molecules as a heterodimer on the surface of nucleated cells and is required for MHC class I protein expression. [00138] CIITA or CIITA or C2TA are used interchangeably herein and with reference to the nucleic acid sequence or protein sequence of class II major histocompatibility complex transactivator; the human gene has accession number NC_000016.10 (range 10866208..10941562), reference GRCh38.p13. The CIITA protein in the nucleus acts as a positive regulator of MHC class II gene transcription and is required for MHC class II protein expression. [00139] MHC or MHC molecule(s) or MHC protein or MHC complex(es), refer to a major histocompatibility complex molecule (or plural), and include e.g., MHC class I and MHC class II molecules. In humans, MHC molecules are referred to as human leukocyte antigen complexes or HLA molecules or HLA protein. The use of terms MHC and HLA are not meant to be limiting; as used herein, the term MHC may be used to refer to human MHC molecules, i.e., HLA molecules. Therefore, the terms MHC and HLA are used interchangeably herein. [00140] HLA-A as used herein in the context of HLA-A protein, refers to the MHC class I protein molecule, which is a heterodimer consisting of a heavy chain (encoded by the HLA-A gene) and a light chain (i.e., beta-2 microglobulin). The terms HLA-A or HLA-A gene, as used herein in the context of nucleic acids refers to the gene encoding the heavy chain of the HLA- A protein molecule. The HLA-A gene is also referred to as HLA class I histocompatibility, A alpha chain; the human gene has accession number NC_000006.12 (29942532..29945870). The HLA-A gene is known to have hundreds of different versions (also referred to as alleles) across the population (and an individual may receive two different alleles of the HLA-A gene). All alleles of HLA-A are encompassed by the terms HLA-A and HLA-A gene. [00141] HLA-B as used herein in the context of nucleic acids refers to the gene encoding the heavy chain of the HLA-B protein molecule. The HLA-B is also referred to as HLA class I histocompatibility, B alpha chain; the human gene has accession number NC_000006.12 (31353875..31357179).
Attorney Docket: 01155-0064-00PCT [00142] HLA-C as used herein in the context of nucleic acids refers to the gene encoding the heavy chain of the HLA-C protein molecule. The HLA-C is also referred to as HLA class I histocompatibility, C alpha chain; the human gene has accession number NC_000006.12 (31268749..31272092). [00143] The term homozygous refers to having two identical alleles of a particular gene. [00144] Any cell type described herein may be used in the editing methods disclosed herein. [00145] In some embodiments, the lipid nucleic acid assembly composition is pretreated with a serum factor before contacting the cell. In some embodiments, the lipid nucleic acid assembly composition is pretreated with a human serum before contacting the cell. In some embodiments, the lipid nucleic acid assembly composition is pretreated with ApoE before contacting the cell. In some embodiments, the lipid nucleic acid assembly composition is pretreated with a recombinant ApoE3 or ApoE4 before contacting the cell. In some embodiments, the cell is serum-starved prior to contact with the lipid nucleic acid assembly composition. [00146] In some embodiments, the methods comprise preincubating a serum factor and the lipid nucleic acid assembly composition for about 30 seconds to overnight. In some embodiments, the preincubation step comprises preincubating a serum factor and the lipid nucleic acid assembly composition for about 1 minute to 1 hour. In some embodiments, it comprises preincubating for about 1-30 minutes. In other embodiments, it comprises preincubating for about 1-10 minutes. Still further embodiments comprise preincubating for about 5 minutes. [00147] In some embodiments, the preincubating step occurs at about 4°C. In some embodiments, the preincubating step occurs at about 25°C. In certain embodiments, the preincubating step occurs at about 37°C. The preincubating step may comprise a buffer such as sodium bicarbonate or HEPES. [00148] The terms “genome editing” and “gene editing” are used interchangeably herein. The terms “genome editing tool” and “gene editing tool” are also used interchangeably herein. The terms “nucleic acid genome editing tool” and “genome editing tool” may also be used interchangeably herein. [00149] In some embodiments, the lipid nucleic acid assembly composition is pretreated with a serum factor before contacting the cell. In some embodiments, the lipid nucleic acid assembly composition is pretreated with a human serum before contacting the cell. In some embodiments, the lipid nucleic acid assembly composition is pretreated with a serum replacement, e.g., a commercially available serum replacement, preferably wherein the serum
Attorney Docket: 01155-0064-00PCT replacement is appropriate for ex vivo use. In some embodiments, the lipid nucleic acid assembly composition is pretreated with ApoE before contacting the cell. In some embodiments, the lipid nucleic acid assembly composition is pretreated with a recombinant ApoE3 or ApoE4 before contacting the cell. In some embodiments, the cell is serum-starved prior to contact with the lipid nucleic acid assembly composition. [00150] In some embodiments, the editing methods comprise preincubating a serum factor and the lipid nucleic acid assembly composition for about 30 seconds to overnight. In some embodiments, the preincubation step comprises preincubating a serum factor and the lipid nucleic acid assembly composition for about 1 minute to 1 hour. In some embodiments, it comprises preincubating for about 1-30 minutes. In other embodiments, it comprises preincubating for about 1-10 minutes. Still further embodiments comprise preincubating for about 5 minutes. [00151] In some embodiments, the preincubating step occurs at about 4°C. In some embodiments, the preincubating step occurs at about 25°C. In some embodiments, the preincubating step occurs at about 37°C. The preincubating step may comprise a buffer such as sodium bicarbonate or HEPES. [00152] In some embodiments, a lipid nucleic acid assembly composition is provided to an “activated” cell or a “non-activated” cell. An “activated” cell may be useful in the methods disclosed herein and may refer to a cell to which a step necessary to induce the cell into a state that enables gene editing thereof has been performed. Agents for activating cells in vitro are provided herein and are known in the art, particularly for activation of immune cells such as T cells or B cells or of stem cells such as iPSCs. For example, activation of a T cell may comprise inducing the T cell into cell cycling (proliferation), and activation of an iPSC may comprise treating the cell with a proinflammatory cytokine to induce HLA class I upregulation. A “non- activated” cell refers to a cell to which the step necessary to induce the cell into a state that enables gene editing thereof has not been performed. In some embodiments, a “non-activated” T cell may have been stimulated in vivo (e.g., by antigen) while in the body, however said cell may be referred to as non-activated herein if said cell has not been stimulated in vitro in culture. [00153] In some embodiments, a T cell is cultured in culture medium prior to contact with a lipid nucleic acid assembly composition. In some embodiments, the T cell is cultured with one or more proliferative cytokines, for example one or more or all of IL-2, IL-15 and IL-21, and/or one or more agents that provides activation through CD3 and/or CD28.
Attorney Docket: 01155-0064-00PCT [00154] In some embodiments, the T cell is activated prior to contact with a lipid nucleic acid assembly composition, is activated in between contact with lipid nucleic acid assembly compositions, and/or is activated after contact with a lipid nucleic acid assembly composition. [00155] In some embodiments, the T cell is activated by polyclonal activation (or “polyclonal stimulation”) (not antigen-specific stimulation). In some embodiments, the T cell is activated by CD3 stimulation (e.g., providing an anti-CD3 antibody). In some embodiments, the T cell is activated by CD3 and CD28 stimulation (e.g., providing an anti-CD3 antibody and an anti- CD28 antibody). In some embodiments, the T cell is activated using a ready-to-use reagent to activate the T cell (e.g., via CD3/CD28 stimulation). In some embodiments, the T cell is activated by via CD3/CD28 stimulation provided by beads. In some embodiments, the T cell is activated by via CD3/CD28 stimulation wherein one or more components is soluble and/or one or more components is bound to a solid surface (e.g., plate or bead). In some embodiments, the T cell is activated by an antigen-independent mitogen (e.g., a lectin, including e.g., concanavalin A (“ConA”), or PHA). [00156] In some embodiments, one or more cytokines are used for activation of T cells. IL-2 is provided for T cell activation. In some embodiments, the cytokine(s) for activation of T cells is a cytokine that binds to the common gamma chain (γc) receptor. In some embodiments, IL- 2 is provided for T cell activation. In some embodiments, IL-7 is provided for T cell activation. In some embodiments, IL-7 is provided to promote T cell survival. In some embodiments, IL- 15 is provided for T cell activation. In some embodiments, IL-21 is provided for T cell activation. In some embodiments, a combination of cytokines is provided for T cell activation, including e.g., IL-2, IL-7, IL-15, and/or IL-21. [00157] In some embodiments, the T cell is activated by exposing the cell to an antigen (antigen stimulation). A T cell is activated by antigen when the antigen is presented as a peptide in a major histocompatibility complex (“MHC”) molecule (peptide-MHC complex). A cognate antigen may be presented to the T cell by co-culturing the T cell with an antigen-presenting cell (feeder cell) and antigen. In some embodiments, the T cell is activated by co-culture with an antigen-presenting cell that has been pulsed with antigen. In some embodiments, the antigen-presenting cell has been pulsed with a peptide of the antigen. [00158] In some embodiments, the T cell may be activated for 12 to 72 hours. In some embodiments, the T cell may be activated for 12 to 48 hours. In some embodiments, the T cell may be activated for 12 to 24 hours. In some embodiments, the T cell may be activated for 24 to 48 hours. In some embodiments, the T cell may be activated for 24 to 72 hours. In some
Attorney Docket: 01155-0064-00PCT embodiments, the T cell may be activated for 12 hours. In some embodiments, the T cell may be activated for 48 hours. In some embodiments, the T cell may be activated for 72 hours. [00159] In some embodiments, the methods provided herein do not include a selection step. In some embodiments, a selection step is included, and optionally the selection step is a physical sorting step (e.g., FACS or MACS) or a biochemical selection step (e.g., suicide gene, drug resistant selection, or antibody-toxin conjugate selection). [00160] The lipid nucleic acid assembly compositions disclosed herein may be used in multiplex genome editing methods in vitro. The methods overcome existing problems with such methods by reducing toxicities associated with the transfection process itself. The reduced toxicity of each transfection event allows for multiple transactions and thereby multiple genome edits per cell. [00161] In some embodiments, the genome edit comprises any one or more of an insertion, deletion, or substitution of at least one nucleotide in a target sequence. In some embodiments, the genome edit comprises an insertion of 1, 2, 3, 4 or 5 or more nucleotides in a target sequence. In some embodiments, the genome edit comprises a deletion of 1, 2, 3, 4 or 5 or more nucleotides in a target sequence. In other embodiments, the genome edit comprises an insertion of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20 or 25 or more nucleotides in a target sequence. In other embodiments, the genome edit comprises a deletion of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20 or 25 or more nucleotides in a target sequence. In some embodiments, the genome edit comprises an indel, which is generally defined in the art as an insertion or deletion of less than 1000 base pairs (bp). In some embodiments, the genome edit comprises an indel which results in a frameshift mutation in a target sequence. In some embodiments, the genome edit comprises a substitution of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20 or 25 or more nucleotides in a target sequence. In some embodiments, the genome edit comprises one or more of an insertion, deletion, or substitution of nucleotides resulting from the incorporation of a template nucleic acid. In some embodiments, the genome edit comprises an insertion of a donor nucleic acid in a target sequence. In some embodiments, the edit or modification is not transient. [00162] In some embodiments, one or more donor nucleic acids are provided for insertion in a target sequence. In some embodiments, the target sequence for insertion is a safe harbor locus. A safe harbor locus is a site in the genome able to accommodate the integration of an exogenous sequence without causing adverse alterations in the host genome and are known in the art. In some embodiments, the target sequence for insertion is in the β-2 microglobulin (B2M) gene. In some embodiments, the target sequence for insertion is in the class II major histocompatibility complex transactivator (CIITA) gene. In some embodiments, the target
Attorney Docket: 01155-0064-00PCT sequence for insertion is in the TRAC gene. In some embodiments, the target sequence for insertion is in AAVS1. III. Cell Populations and Methods/Uses A. Cell Populations [00163] In some embodiments, the cells for the methods disclosed herein are selected from stem cells (e.g., mesenchymal stem cells; hematopoietic stem cells (HSCs); neural stem cells (NSCs); limbal stem cells (LSCs); induced pluripotent stem cells (iPSCs); ocular stem cells; pluripotent stem cells (PSCs); and embryonic stem cells (ESCs)); mononuclear cells; endothelial progenitor cells (EPCs); tissue-specific primary cells or cells derived therefrom (TSCs), cells for organ or tissue transplantations, and cells for use in ACT therapy. In some embodiments, the cells for the methods disclosed herein are induced pluripotent stem cells (iPSCs). [00164] In some embodiments, the cells for the methods disclosed herein are selected from immune cells, such as lymphocytes (e.g., T cell, B cell, natural killer cell (“NK cell”, and NKT cell, or iNKT cell)), monocytes, macrophages, mast cells, dendritic cells, granulocytes (e.g., neutrophil, eosinophil, and basophil), primary immune cells, CD3+ cells, CD4+ cells, CD8+ T cells, regulatory T cells (Tregs), B cells, NK cells, and dendritic cells (DC)). In some embodiments, the immune cells are selected from peripheral blood mononuclear cell (PBMC), a lymphocyte, a T cell, optionally a CD4+ cell, a CD8+ cell, a memory T cell, a naïve T cell, a stem-cell memory T cell; or a B cell, optionally a memory B cell, a naïve B cell; and a primary cell. In some embodiments, the immune cells are T cells. In some embodiments, the immune cells are T cells selected from tumor infiltrating lymphocytes (TILs), T cells expressing an alpha-beta TCR, T cells expressing a gamma-delta TCR, a regulatory T cells (Treg), memory T cells, and early stem cell memory T cells (Tscm, CD27+/CD45+). [00165] In some embodiments, the immune cells are isolated from human donor PBMCs or leukopaks before editing. In some embodiments, the immune cells are derived from a progenitor cell. [00166] In some embodiments, the cells are non-activated cells. In some embodiments, the cells are activated cells. In some embodiments, the cells have already been activated prior to performance of any one of the methods disclosed herein. [00167] In some embodiments, the cells have reduced or eliminated surface expression of HLA-A. In some embodiments, no more than 4%, no more than 5%, no more than 6%, no more than 7%, no more than 8%, or no more than 9% of the cells are positive for HLA-A surface
Attorney Docket: 01155-0064-00PCT expression. In some embodiments, no more than 4% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 5% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 6% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 7% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 8% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 9% of the cells are positive for HLA-A surface expression. In some embodiments, the HLA-A surface expression is measured by flow cytometry. In some embodiments, the cells are homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the cells edited by the methods disclosed herein have reduced or eliminated surface expression of HLA-A, and the cells are homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the cells edited by the methods disclosed herein have reduced or eliminated surface expression of HLA-A and HLA-B, and the cells are homozygous for HLA-C. In some embodiments, the cells are HLA-H1 positive and are homozygous or heterozygous for HLA-H1. In some embodiments, the cells are homozygous or heterozygous for HLA-H1. [00168] In some embodiments, provided herein are a cell population made by or obtainable by any one of the methods disclosed herein. [00169] In some embodiments, a rate of Chr6p21.3 deletion in the cell population is no more than 2 per 200 metaphase spreads. In some embodiments, a copy number variation (CNV) of the short arm of human chromosome 6 in the cell population is no less than 1.90. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.91. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.92. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.93. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.94. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.95. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.96. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.97. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.98. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 1.99. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 2.00. In some embodiments, a CNV of the short arm of human chromosome 6 in the cell population is no less than 2.01. In some embodiments, a CNV of the
Attorney Docket: 01155-0064-00PCT short arm of human chromosome 6 in the cell population is no less than 2.02. In some embodiments, a rate of chromosome deletion and/or a CNV of the short arm of human chromosome 6 in a cell population made by or obtainable by any one of the methods disclosed herein is no more than a background rate of chromosome deletion and/or a background CNV of the short arm of human chromosome 6, respectively, in a same cell population on which the method has not been performed. In some embodiments, a rate of Chr6p21.3 deletion and/or a CNV of the short arm of human chromosome 6 in a cell population made by or obtainable by any one of the methods disclosed herein is no more than a background rate of Chr6p21.3 deletion and/or a background CNV of the short arm of human chromosome 6, respectively, in a same cell population on which the method has not been performed. In some embodiments, the rate of chromosome deletion is measured by a karyotype analysis. In some embodiments, the rate of Chr6p21.3 deletion is measured by a karyotype analysis. In some embodiments, the CNV is measured using a NEDD9 gene as a marker for the distal portion of the short arm of human chromosome 6 and using a MRPL18 gene as a reference control on the long arm of human chromosome 6. In some embodiments, the CNV is measured by droplet digital PCR (ddPCR). [00170] In some embodiments, the cells of the cell population are for use in a method of therapy or a pharmaceutical composition. In some embodiments, the method of therapy or the pharmaceutical composition are for treatment of cancer or autoimmune therapy. In some embodiments, the method of therapy or pharmaceutical composition are for adoptive cell transfer therapy. In some embodiments, the cells of the cell population are for transfer into a human subject. In some embodiments, as a method of creating a cell bank, the cells of the cell population are transferred into a cell bank. [00171] In some embodiments, at least 95% of the cells in the cell population comprise a genome edit of an endogenous TCR sequence. In some embodiments, at least 96% of the cells in the cell population comprise a genome edit of an endogenous TCR sequence. In some embodiments, at least 97% of the cells in the cell population comprise a genome edit of an endogenous TCR sequence. In some embodiments, at least 98% of the cells in the cell population comprise a genome edit of an endogenous TCR sequence. In some embodiments, at least 99% of the cells in the cell population comprise a genome edit of an endogenous TCR sequence. [00172] In some embodiments, the cell population comprises edited cells with a genome edit comprising an insertion of an exogenous nucleic acid sequence coding for a targeting ligand or an alternative antigen binding moiety wherein at least 70% of the cells of the cell
Attorney Docket: 01155-0064-00PCT population comprise an insertion of an exogenous nucleic acid into a target sequence. In some embodiments, the cell population comprises edited cells with a genome edit comprising an insertion of an exogenous nucleic acid sequence coding for a targeting ligand or an alternative antigen binding moiety wherein at least 80% of the cells of the cell population comprise an insertion of an exogenous nucleic acid into a target sequence. In some embodiments, the cell population comprises edited cells with a genome edit comprising an insertion of an exogenous nucleic acid coding for a targeting ligand or an alternative antigen binding moiety wherein at least 90% of the cells of the cell population comprise an insertion of an exogenous nucleic acid into a target sequence. In some embodiments, the cell population comprises edited cells with a genome edit comprising an insertion of an exogenous nucleic acid coding for a targeting ligand or an alternative antigen binding moiety wherein at least 95% of the cells of the cell population comprise an insertion of an exogenous nucleic acid into a target sequence. [00173] In some embodiments, the cell population comprises edited T cells, wherein at least 30%, 40%, 50%, 55%, 60%, or 65% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). In some embodiments, the cell population comprises edited T cells, wherein at least 30% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). In some embodiments, the cell population comprises edited T cells, wherein at least 40% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). In some embodiments, the cell population comprises edited T cells, wherein at least 50% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). In some embodiments, the cell population comprises edited T cells, wherein at least 55% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). In some embodiments, the cell population comprises edited T cells, wherein at least 60% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). In some embodiments, the cell population comprises edited T cells, wherein at least 65% of the cells of the cell population have a memory phenotype (CD27+, CD45RA+). [00174] In some embodiments, the cell population comprising edited cells comprises cells with reduced or eliminated surface expression of MHC class I and/or MHC class II. In some embodiments, the cell population comprising edited cells comprises cells with reduced or eliminated surface expression of both MHC class I and MHC class II. In some embodiments, the cell population comprising edited cells comprises cells with reduced or eliminated surface expression of HLA-A. In some embodiments, no more than 4%, no more than 5%, no more than 6%, no more than 7%, no more than 8%, or no more than 9% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 4% of the cells are positive
Attorney Docket: 01155-0064-00PCT for HLA-A surface expression. In some embodiments, no more than 5% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 6% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 7% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 8% of the cells are positive for HLA-A surface expression. In some embodiments, no more than 9% of the cells are positive for HLA-A surface expression. In some embodiments, the HLA-A surface expression is measured by flow cytometry. In some embodiments, the cells in the cell population comprise a genetic modification in a HLA-A gene. In some embodiments, the genetic modification in the HLA-A gene comprises at least one nucleotide within the genomic coordinates chr6:29942864-29942884. In some embodiments, the cell population comprising edited cells comprises cells with reduced or eliminated surface expression of HLA-A, and the cells are homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the cell population comprising edited cells comprises cells with reduced or eliminated surface expression of HLA-A and HLA-B, and the cells are homozygous for HLA-C. [00175] In some embodiments, the cell population comprising edited T cells comprises cells with reduced or eliminated surface expression of MHC class I and/or MHC class II. In some embodiments, the cell population comprising edited T cells comprises cells with reduced or eliminated surface expression of both MHC class I and MHC class II. In some embodiments, the cell population comprising edited T cells comprises cells with reduced or eliminated surface expression of HLA-A, and the cells are homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the cell population comprising edited T cells comprises cells with reduced or eliminated surface expression of HLA-A and HLA-B, and the cells are homozygous for HLA-C. [00176] In some embodiments, a population of cells is produced according to the provided methods. In some embodiments, at least 50% or more of the cells in the population comprises more than one genome edit. In some embodiments, at least 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% (i.e., all cells as determined by the method of detection) of the cells in the population comprises more than one genome edit. In some embodiments, a method disclosed herein results in at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, preferably at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90% or at least 95% of the cells having at least two genome edits. In other embodiments, a method disclosed herein, results in at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, preferably at least 30%, at least 35%, at least
Attorney Docket: 01155-0064-00PCT 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90% or at least 95% of the cells having 2, 3, 4, 5, 6, 7, or 8 genome edits. In some embodiments, a method disclosed herein results in about 5% to about 100%, about 10% to about 50%, about 20 to about 100%, about 20 to about 80%, about 40 to about 100%, or about 40 to about 80% of every cell in a population having at least two genome edits. In some embodiments, the cells have not undergone a selection process, e.g., FACS or a biochemical selection process, at the completion of editing to enrich the population for edited cells. [00177] In some embodiments, the methods disclosed herein produce expanded cells in vitro with increased survival. In embodiments, the improved survival rate may be compared to cells treated with electroporation processes. In some embodiments, the cell survival rate of an expanded cell is at least 70%, 80%, 90%, or 95%. [00178] In some embodiments, at least 70%, 80%, 90%, or 95% of the cells on which the methods disclosed herein are performed are viable 24 hours after contacting the population of cells with an LNP composition. In some embodiments, at least 70% of the cells on which the methods disclosed herein are performed are viable 24 hours after contacting the population of cells with an LNP composition. In some embodiments, at least 80% of the cells on which the methods disclosed herein are performed are viable 24 hours after contacting the population of cells with an LNP composition. In some embodiments, at least 90% of the cells on which the methods disclosed herein are performed are viable 24 hours after contacting the population of cells with an LNP composition. In some embodiments, at least 95% of the cells on which the methods disclosed herein are performed are viable 24 hours after contacting the population of cells with an LNP composition. [00179] In some embodiments, the methods disclosed herein produced cells in vitro with low toxicity. For example, in embodiments, the resultant cells of the disclosed methods have less than 2%, 1%, 0.5%, 0.2%, 0.1% translocations, including e.g., target-target translocations, and/or off-target translocations. In some embodiments, the resultant cells of the disclosed method have less than 1%, 0.5%, 0.2%, 0.1% target-target translocations. In some embodiments, the resultant cells of the disclosed methods no measurable translocations, including e.g., target-target translocations, and/or off-target translocations. In some embodiments, the resultant cells have no measurable reciprocal translocations as determined, for example, using the methods provided herein. In some embodiments, the resultant cells have no measurable complex translocations as determined, for example, using the methods provided herein. In some embodiments, the resultant cells have no measurable off-target translocations
Attorney Docket: 01155-0064-00PCT as determined, for example, using the methods provided herein. In some embodiments, the resultant cells have less than 2 times the background level of reciprocal translocations, complex translocations, or off-target translocations, as determined, for example, using the methods provided herein. [00180] In some embodiments, the methods disclosed herein produce cells with high editing efficiency. A particular advantage of the disclosed methods are the high editing rates observed in cells having multiple genome edits. For example, in some embodiments, the percent editing efficiency is at least 60%, 70%, 80%, 90%, or 95% at each target site. [00181] It is understood that the number of cells in a population needed for any particular use depends, for example, on the type of cell and the intended use of the cell. The number of cells to be edited also depends on the ability to proliferate the cells after editing. It is also understood that the level of editing required, or the level of knockdown required, depends, at least in part, on the particular edit being made and the intended use of the cell population. For example, a population of B cells with genome editing, e.g., of 30% or less, 40% or less, 50% or less, may be useful in a protein expression system. For example, higher levels of knockdown are required of endogenous T cell receptor (TCR) on the surface of a T cell for transplantation into a subject, as low levels of endogenous TCR on the surface of the T cell can result in a severe adverse reaction when transplanted into a subject. Therefore, T cells expressing an endogenous TCR should be present in as low levels as possible in a population of T cells for transplantation purposes. However, editing of a T cell to produce a cytokine or other secreted factor, even for use in transplantation, may not require as high levels of editing as would be required for the endogenous TCR in a population of T cells for transplantation. [00182] Exemplary edited cell population sizes are provided below. It is understood that the number of edited cells required for any particular indication may vary, e.g., therapeutic methods, may vary. Also, larger numbers of cells may be desirable for cell populations for use in allogenic therapies than for autologous therapies. [00183] In certain embodiments, the population of cells comprising edited cells is a population of T cells. In certain embodiments the population of T cells comprises 1 x 10^9 edited T cells with multiple, i.e., at least 2, edits. In certain embodiments the population of T cells comprises 5 x 10^9 edited T cells with at least a single edit. In certain embodiments, the population of T cells comprises 1-10 x 10^9 edited T cells and is useful for TCR-T cell therapy. In certain embodiments, the population of T cells comprises 1 x 10^8 edited T cells and is useful for CAR-T therapy.
Attorney Docket: 01155-0064-00PCT [00184] In certain embodiments, the population of cells comprising edited cells is a population of B cells. In certain embodiments, the population of B cells comprises 1-5 x 10e8 edited B cells with at least a single edit, preferably comprising edited B cells with multiple edits. [00185] In certain embodiments, the population of cells comprising edited cells is a population of NK cells. In certain embodiments, the population of NK cells comprises 3 x 10^9 NK edited NK cells with at least a single edit. In certain embodiments, the population of NK cells comprises at least 5 x 10^8 edited NK cells with multiple edits. In certain embodiments, the population of NK cells comprises 1 x 10^8 to 9 x 10^9 edited NK cells for use in therapy. [00186] In certain embodiments, the population of cells comprising edited cells is a population of monocytes or macrophages. In certain embodiments, the population of monocytes or macrophages comprising edited cells comprises at least 1 x 10^9 monocytes or macrophages having at least a single edit, or at least 2 x 10^8 monocytes or macrophages with multiple edits. [00187] In certain embodiments, the population of cells comprising edited cells are dendritic cells. In certain embodiments, the population of dendritic cells comprises 5 x 10^6 to 5 x 10^7 edited dendritic cells. [00188] In some embodiments, the genome editing methods to T cells in vitro have produced high editing efficiency at multiple target sites. In some embodiments, an engineered T cell is produced wherein the endogenous TCR is knocked out. In some embodiments, an engineered T cell is produced wherein expression of the endogenous TCR is reduced. In some embodiments, an engineered T cell is produced wherein three genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein four genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein five genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein six genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein seven genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein eight genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein nine genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein ten genes have reduced expression and/or are knocked out. In some embodiments, an engineered T cell is produced wherein eleven genes have reduced expression and/or are knocked out.
Attorney Docket: 01155-0064-00PCT [00189] In some embodiments, an engineered T cell is produced wherein the endogenous TCR is knocked out and a transgenic TCR is inserted and expressed. In some embodiments, the engineered T cell is a primary human T cell. In some embodiments, the tgTCR targets Wilms’ Tumor 1 (WT1). In some embodiments, the WT1 tgTCR is inserted into a high proportion of T cells (e.g., greater than 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or 95%) using the disclosed lipid nucleic acid assembly composition. [00190] In some embodiments, the T cells produced by the disclosed methods have increased production of cytokines. In some embodiments, the increase in production of cytokines may be compared to T cells treated with electroporation processes. For example, in some embodiments, the genetically engineered T cells produced increased levels of IL-2. In some embodiments, the genetically engineered T cells produced increased levels of IFNγ. In some embodiments, the genetically engineered T cells produced increased levels of TNFα. Cytokine levels may be determined by standard methods, including e.g., ELISA, intracellular flow cytometry staining. [00191] In some embodiments, the T cells produced by the disclosed methods demonstrate continued proliferation with repeat stimulation. For example, the T cells may proliferate following repeat stimulation in in vitro culture with an agent used to stimulate a T cell. In some embodiments, the T cell may be stimulated and proliferate in response to repeat stimulation with the cognate antigen for the T cell’s TCR (e.g., peptide-MHC complexes on a cell that is co-cultured with the T cell). In some embodiments, the T cell may be stimulated and proliferate in response to repeat polyclonal stimulation. In some embodiments, the repeat stimulation is at least twice, three times, four times, five times, or more. In some embodiments, a proliferating cell is expanded to form a population of cells that comprise the genetic modification. [00192] In some embodiments, the T cells produced by the disclosed methods demonstrate increased expansion. In some embodiments, the increase in expansion may be compared to T cells treated with electroporation processes. Expansion may be evaluated by cell count, proliferation, or other standard methods for measuring expansion of T cells. [00193] In some embodiments, the T cells produced by the disclosed methods exhibit a memory T cell phenotype. In some embodiments, the T cell memory phenotype referred to early stem-cell memory T cells (or “Tscm”) are particularly advantageous and are produced by the disclosed methods. In some embodiments, a genetically engineered T cell has the Tscm phenotype (CD27+, CD45RA+). [00194] In some embodiments, the engineered cell (e.g., T cell) produced by the disclosed method has reduced or eliminated surface expression of MHC class I and/or MHC class II. In
Attorney Docket: 01155-0064-00PCT some embodiments, the engineered cell has reduced or eliminated surface expression of both MHC class I and MHC class II. In some embodiments, the engineered cell has reduced or eliminated surface expression of HLA-A, and the cell is homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the engineered cell has reduced or eliminated surface expression of HLA-A and HLA-B, and the cell is homozygous for HLA-C. [00195] In some embodiments, the engineered T cell produced by the disclosed methods has reduced or eliminated surface expression of MHC class I and/or MHC class II. In some embodiments, the engineered cell has reduced or eliminated surface expression of both MHC class I and MHC class II. In some embodiments, the engineered cell has reduced or eliminated surface expression of HLA-A, and the cell is homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the engineered cell has reduced or eliminated surface expression of HLA-A and HLA-B, and the cell is homozygous for HLA-C. B. Methods/Uses for Treating Disorders [00196] The cell and/or population of cells made by or obtainable by any one of the methods provided herein may be used in methods of treating a variety of diseases and disorders. [00197] In some embodiments, the disclosure provides a method of providing an immunotherapy in a subject, the method including administering to the subject an effective amount of a cell (e.g., a population of cells) as described herein, for example, a cell of any of the aforementioned cell aspects and embodiments. [00198] In some embodiments of the methods, the method includes administering a lymphodepleting agent or immunosuppressant prior to administering to the subject an effective amount of the cell (e.g., a population of cells) as described herein, for example, a cell of any of the aforementioned cell aspects and embodiments. In another aspect, the disclosure provides a method of preparing cells (e.g., a population of cells). [00199] Immunotherapy is the treatment of disease by activating or suppressing the immune system. Immunotherapies designed to elicit or amplify an immune response are classified as activation immunotherapies. Cell-based immunotherapies have been demonstrated to be effective in the treatment of some cancers. Immune effector cells such as lymphocytes, macrophages, dendritic cells, natural killer cells, cytotoxic T lymphocytes (CTLs) can be programmed to act in response to abnormal antigens expressed on the surface of tumor cells. Thus, cancer immunotherapy allows components of the immune system to destroy tumors or other cancerous cells. Cell-based immunotherapies have also been demonstrated to be effective in the treatment of autoimmune diseases or transplant rejection. Immune effector cells such as
Attorney Docket: 01155-0064-00PCT regulatory T cells (Tregs) or mesenchymal stem cells can be programmed to act in response to autoantigens or transplant antigens expressed on the surface of normal tissues. [00200] In some embodiments, the disclosure provides a population of cells or a method of preparing cells (e.g., a population of cells). The population of cells may be used for immunotherapy. [00201] Cells of the disclosure are suitable for further engineering, e.g., by introduction of further edited, or modified genes or alleles. In some embodiments, the polypeptide is a wild- type or variant TCR. Cells of the disclosure may also be suitable for further engineering by introduction of a heterologous sequence coding for an alternative antigen binding moiety, e.g., by introduction of a heterologous sequence coding for an alternative (non-endogenous) TCR, e.g., a chimeric antigen receptor (CAR) engineered to target a specific protein. CARs are also known as chimeric immunoreceptors, chimeric T cell receptors or artificial T cell receptors. [00202] In some embodiments, the disclosure provides a method of treating a subject in need thereof that includes administering cells (e.g., a population of cells), e.g., cells prepared by a method described herein, for example, a method of any of the aforementioned aspects and embodiments. [00203] In some embodiments, the population of cells or cells produced by the disclosed methods can be used to treat cancer, infectious diseases, inflammatory diseases, autoimmune diseases, cardiovascular diseases, neurological diseases, ophthalmologic diseases, renal diseases, liver diseases, musculoskeletal diseases, red blood cell diseases, or transplant rejections. [00204] In some embodiments, the cancer is lymphoma, breast cancer, lung cancer, multiple myeloma, leukemia, liver cancer, urinary tract cancer, kidney cancer, bladder cancer, melanoma, colorectal cancer, pancreatic cancer, epithelial malignancies, mesothelioma, oropharyngeal cancer, cervical cancer, uterine cancer, ovarian cancer, anogenital cancer, or brain cancer. In some embodiments, the lymphoma is non-Hodgkin’s lymphoma, including diffuse large B cell lymphoma (DLBCL), aggressive B cell lymphoma, or high-grade B cell lymphoma, or mantle cell lymphoma. In some embodiments, the breast cancer is a triple negative breast cancer. In some embodiments, the lung cancer is non-small cell lung cancer (NSCLC) or small cell lung cancer (SCLC). In some embodiments, the leukemia is acute lymphoblastic leukemia or acute myeloid leukemia. In some embodiments, the cancer is a solid tumor. [00205] In some embodiments, the infectious disease is caused by human immunodeficiency virus (HIV), Hepatitis A virus, Hepatitis C Virus, Hepatitis B Virus, Human
Attorney Docket: 01155-0064-00PCT Cytomegalovirus (CMV), Epstein-Barr virus, human papillomavirus, Mycobacterium tuberculosis, a human coronavirus, or invasive Aspergillus fumigatus. In some embodiments, the infectious disease is acquired immunodeficiency syndrome (AIDS), hepatitis A, hepatitis B, hepatitis C, tuberculosis, severe acute respiratory syndrome (SARS), middle east respiratory syndrome (MERS), or coronavirus disease 2019 (COVID-19). In some embodiments, the tuberculosis is multidrug-resistant (MDR) tuberculosis or extensively drug-resistant (XDR) tuberculosis. In some embodiments, the human coronavirus is middle east respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS- CoV), or severe acute respiratory syndrome coronavirus 2 (SARS-CoV2). In some embodiments, infectious disease is a human papillomavirus-positive cancer, such as uterine cancer, cervical cancer, or oropharyngeal cancer. [00206] In some embodiments, the inflammatory disease is allergy, asthma, celiac disease, glomerulonephritis, inflammatory bowel disease, gout, rheumatoid arthritis (RA), myositis, scleroderma, ankylosing spondylitis (AS), antiphospholipid antibody syndrome (APS), systemic lupus erythematosus (SLE), Sjogren’s syndrome, rheumatic heart disease, chronic obstructive pulmonary disease (COPD), or transplant rejection. [00207] In some embodiments, the autoimmune disease is Type 1 diabetes, multiple sclerosis, Crohn’s diseases, ulcerative colitis, autoimmune thyroid disease, rheumatoid arthritis (RA), inflammatory bowel disease, antiphospholipid antibody syndrome (APS), Sjogren’s syndrome, scleroderma, psoriasis, psoriatic arthritis, Guillain-Barre syndrome, Addison’s disease, Graves’ disease, Hashimoto’s thyroiditis, Myasthenia gravis, autoimmune vasculitis, autoimmune uveitis, autoimmune hepatitis, pernicious anemia, celiac disease, or systemic lupus erythematosus (SLE). [00208] In some embodiments, the cardiovascular disease is ischemic heart disease, coronary heart disease, aorta disease, Marfan syndrome, congenital heart disease, heart valve disease, pericardial disease, rheumatic heart disease, peripheral arterial disease, or stroke. [00209] In some embodiments, the neurological disease is Parkinson’s disease, amyotrophic lateral sclerosis, stroke, spinal cord injury, Alzheimer’s disease, age-related macular degeneration, traumatic brain injury, multiple sclerosis, Huntington’s disease, muscular dystrophy, or Guillain-Barre syndrome. [00210] In some embodiments, the ophthalmologic disease is glaucoma, retinopathy, macular degeneration, or cytomegalovirus (CMV) retinitis. In some embodiments, the ophthalmologic disease is a retinal disease. In some embodiments, the ophthalmologic disease is mediated by VEGF.
Attorney Docket: 01155-0064-00PCT [00211] In some embodiments, the engineered cells produced by the disclosed methods can be used as a cell therapy comprising an autologous cell therapy. In some embodiments, the engineered cells can be used as a cell therapy comprising an allogeneic stem cell therapy. In some embodiments, the cell therapy comprises induced pluripotent stem cells (iPSCs). iPSCs may be induced to differentiate into other cell types including, e.g., beta islet cells, neurons, and blood cells. In some embodiments, the cell therapy comprises hematopoietic stem cells. In some embodiments, the stem cells comprise mesenchymal stem cells that can develop into bone, cartilage, muscle, and fat cells. In some embodiments, the stem cells comprise ocular stem cells. In some embodiments, the allogeneic stem cell transplant comprises allogeneic bone marrow transplant. In some embodiments, the stem cells comprise pluripotent stem cells (PSCs). In some embodiments, the stem cells comprise induced embryonic stem cells (ESCs). [00212] In some embodiments, the cell therapy is a transgenic T cell therapy. In some embodiments, the cell therapy comprises a Wilms’ Tumor 1 (WT1) targeting transgenic T cell. In some embodiments, the cell therapy comprises a targeting receptor or a donor nucleic acid encoding a targeting receptor of a commercially available T cell therapy, such as a CAR T cell therapy. There are a number of targeting receptors currently approved for cell therapy. The cells and methods provided herein can be used with these known constructs. Commercially approved cell products that include targeting receptor constructs for use as cell therapies include e.g., Kymriah® (tisagenlecleucel); Yescarta® (axicabtagene ciloleucel); Tecartus™ (brexucabtagene autoleucel); Tabelecleucel (Tab-cel®); Abecma® (idecabtagene vicleucel); Carvykti® (ciltacabtagene autoleucel); Viralym-M (ALVR105); and Viralym-C. C. Exemplary Cell Types [00213] In some embodiments, the cell is an immune cell. As used herein, “immune cell” refers to a cell of the immune system, including e.g., a lymphocyte (e.g., T cell, B cell, natural killer cell (“NK cell”, and NKT cell, or iNKT cell)), monocyte, macrophage, mast cell, dendritic cell, or granulocyte (e.g., neutrophil, eosinophil, and basophil). In some embodiments, the cell is a primary immune cell. In some embodiments, the immune system cell may be selected from CD3
+, CD4
+ and CD8
+ T cells, regulatory T cells (Tregs), B cells, NK cells, and dendritic cells (DC). In some embodiments, the immune cell is allogeneic. [00214] In some embodiments, the cell is a lymphocyte. In some embodiments, the cell is an adaptive immune cell. In some embodiments, the cell is a T cell. In some embodiments, the cell is a B cell. In some embodiments, the cell is a NK cell.
Attorney Docket: 01155-0064-00PCT [00215] As used herein, a T cell can be defined as a cell that expresses a T cell receptor (“TCR” or “αβ TCR” or “γδ TCR”), however in some embodiments, the TCR of a T cell may be genetically modified to reduce its expression (e.g., by genetic modification to the TRAC or TRBC genes), therefore expression of the protein CD3 may be used as a marker to identify a T cell by standard flow cytometry methods. CD3 is a multi-subunit signaling complex that associates with the TCR. Thus, a T cell may be referred to as CD3+. In some embodiments, a T cell is a cell that expresses a CD3+ marker and either a CD4+ or CD8+ marker. [00216] In some embodiments, the T cell expresses the glycoprotein CD8 and therefore is CD8+ by standard flow cytometry methods and may be referred to as a “cytotoxic” T cell. In some embodiments, the T cell expresses the glycoprotein CD4 and therefore is CD4+ by standard flow cytometry methods and may be referred to as a “helper” T cell. CD4+ T cells can differentiate into subsets and may be referred to as a Th1 cell, Th2 cell, Th9 cell, Th17 cell, Th22 cell, T regulatory (“Treg”) cell, or T follicular helper cells (“Tfh”). Each CD4+ subset releases specific cytokines that can have either proinflammatory or anti-inflammatory functions, survival or protective functions. A T cell may be isolated from a subject by CD4+ or CD8+ selection methods. [00217] In some embodiments, the T cell is a memory T cell. In the body, a memory T cell has encountered antigen. A memory T cell can be located in the secondary lymphoid organs (central memory T cells) or in recently infected tissue (effector memory T cells). A memory T cell may be a CD8+ T cell. A memory T cell may be a CD4+ T cell. [00218] As used herein, a “central memory T cell” can be defined as an antigen-experienced T cell, and for example, may express CD62L and CD45RO. A central memory T cell may be detected as CD62L+ and CD45RO+ by central memory T cells also express CCR7, therefore may be detected as CCR7+ by standard flow cytometry methods. [00219] As used herein, an “early stem-cell memory T cell” (or “Tscm”) can be defined as a T cell that expresses CD27 and CD45RA, and therefore is CD27+ and CD45RA+ by standard flow cytometry methods. A Tscm does not express the CD45 isoform CD45RO, therefore a Tscm will further be CD45RO- if stained for this isoform by standard flow cytometry methods. A CD45RO- CD27+ cell is therefore also an early stem-cell memory T cell. Tscm cells further express CD62L and CCR7, therefore may be detected as CD62L+ and CCR7+ by standard flow cytometry methods. Early stem-cell memory T cells have been shown to correlate with increased persistence and therapeutic efficacy of cell therapy products. [00220] In some embodiments, the cell is a B cell. As used herein, a “B cell” can be defined as a cell that expresses CD19 and/or CD20, and/or B cell mature antigen (“BCMA”), and
Attorney Docket: 01155-0064-00PCT therefore a B cell is CD19+, and/or CD20+, and/or BCMA+ by standard flow cytometry methods. A B cell is further negative for CD3 and CD56 by standard flow cytometry methods. The B cell may be a plasma cell. The B cell may be a memory B cell. The B cell may be a naïve B cell. The B cell may be IgM+ or has a class-switched B cell receptor (e.g., IgG+, or IgA+). [00221] In some embodiments, the cell is a mononuclear cell, such as from bone marrow or peripheral blood. In some embodiments, the cell is a peripheral blood mononuclear cell (“PBMC”). In some embodiments, the cell is a PBMC, e.g., a lymphocyte or monocyte. In some embodiments, the cell is a peripheral blood lymphocyte (“PBL”). [00222] Cells used in ACT therapy are included, such as mesenchymal stem cells (e.g., isolated from bone marrow (BM), peripheral blood (PB), placenta, umbilical cord (UC) or adipose); hematopoietic stem cells (HSCs; e.g.. isolated from BM); mononuclear cells (e.g., isolated from BM or PB); endothelial progenitor cells (EPCs; isolated from BM, PB, and UC); neural stem cells (NSCs); limbal stem cells (LSCs); or tissue-specific primary cells or cells derived therefrom (TSCs). Cells used in ACT therapy further include induced pluripotent stem cells (iPSCs; see e.g., Mahla, International J. Cell Biol. 2016 (Article ID 6940283): 1-24 (2016)) that may be induced to differentiate into other cell types including e.g., islet cells, neurons, and blood cells; ocular stem cells; pluripotent stem cells (PSCs); embryonic stem cells (ESCs); cells for organ or tissue transplantations such as islet cells, cardiomyocytes, thyroid cells, thymocytes, neuronal cells, skin cells, retinal cells, chondrocytes, myocytes, and keratinocytes. [00223] In some embodiments, the cell is a human cell, such as a cell from a subject. In some embodiments, the cell is isolated from a human subject. In some embodiments, the cell is isolated from a patient. In some embodiments, the cell is isolated from a donor. In some embodiments, the cell is isolated from human donor PBMCs or leukopaks. In some embodiments, the cell is from a subject with a condition, disorder, or disease. In some embodiments, the cell is from a human donor with Epstein Barr Virus (“EBV”). [00224] In some embodiments, the cell is homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the cell contains a genetic modification in the HLA-A gene and is homozygous for HLA-B and homozygous for HLA-C. In some embodiments, the cell contains a genetic modification in the HLA-A gene and a genetic modification in the HLA-B gene, and is homozygous for HLA-C. [00225] In some embodiments, the methods are carried out ex vivo. As used herein, “ex vivo” refers to an in vitro method wherein the cell is capable of being transferred into a subject, e.g.
Attorney Docket: 01155-0064-00PCT as an ACT therapy. In some embodiments, an ex vivo method is an in vitro method involving an ACT therapy cell or cell population. [00226] In some embodiments, the cell is maintained in culture. In some embodiments, the cell is transplanted into a patient. In some embodiments, the cell is removed from a subject, genetically modified ex vivo, and then administered back to the same patient. In some embodiments, the cell is removed from a subject, genetically modified ex vivo, and then administered to a subject other than the subject from which it was removed. In some embodiments, the cell is cultured, expanded, and/or proliferated ex vivo either before or after gene editing. In some embodiments, the cell is cultured, expanded, and/or proliferated ex vivo before gene editing. In some embodiments, the cell is cultured, expanded, and/or proliferated ex vivo after gene editing. [00227] In some embodiments, the cell is from a cell line. In some embodiments, the cell line is derived from a human subject. In some embodiments, the cell line is a lymphoblastoid cell line (“LCL”). The cell may be cryopreserved and thawed. The cell may not have been previously cryopreserved. [00228] In some embodiments, the cell is from a cell bank. In some embodiments, the cell is genetically modified and then transferred into a cell bank. In some embodiments the cell is removed from a subject, genetically modified ex vivo, and transferred into a cell bank. In some embodiments, a genetically modified population of cells is transferred into a cell bank. In some embodiments, a genetically modified population of cells is transferred into a cell bank. In some embodiments, a genetically modified population of cells comprising a first and second subpopulations, wherein the first and second sub-populations have at least one common genetic modification and at least one different genetic modification are transferred into a cell bank. IV. Exemplary Genome Editing Tools [00229] In some embodiments, the lipid nucleic acid assembly comprises a genome editing tool or a nucleic acid encoding the same. [00230] As used herein, the term “genome editing tool” (or “gene editing tool”) is any component of “genome editing system” (or “gene editing system”) necessary or helpful for producing an edit in the genome of a cell. In some embodiments, the present disclosure provides for methods of delivering genome editing tools of a genome editing system (for example a zinc finger nuclease system, a TALEN system, a meganuclease system or a CRISPR/Cas system) to a cell (or population of cells). Genome editing tools include, for example, nucleases capable of making single or double strand break in the DNA or RNA of a
Attorney Docket: 01155-0064-00PCT cell, e.g., in the genome of a cell. The genome editing tools, e.g. nucleases, may optionally modify the genome of a cell without cleaving the nucleic acid, or nickases. A genome editing nuclease or nickase may be encoded by an mRNA. Such nucleases include, for example, RNA- guided DNA binding agents, and CRISPR/Cas components. Genome editing tools include fusion proteins, including e.g., a nickase fused to an effector domain such as an editor domain. Genome editing tools include any item necessary or helpful for accomplishing the goal of a genome edit, such as, for example, guide RNA, sgRNA, dgRNA, donor nucleic acid, and the like. [00231] Various suitable gene editing systems comprising genome editing tools for delivery with the lipid nucleic acid assembly compositions are described herein, including but not limited to the CRISPR/Cas system; zinc finger nuclease (ZFN) system; and the transcription activator-like effector nuclease (TALEN) system. Generally, the gene editing systems involve the use of engineered cleavage systems to induce a double strand break (DSB) or a nick (e.g., a single strand break, or SSB) in a target DNA sequence. Cleavage or nicking can occur through the use of specific nucleases such as engineered ZFN, TALENs, or using the CRISPR/Cas system with an engineered guide RNA to guide specific cleavage or nicking of a target DNA sequence. Further, targeted nucleases are being developed based on the Argonaute system (e.g., from T. thermophilus, known as ‘TtAgo’, see Swarts et al (2014) Nature 507(7491): 258-261), which also may have the potential for uses in genome editing and gene therapy. A. CRISPR/Cas Genome Editing Tools [00232] In some embodiments, the genome editing tool is a component of a CRISPR/Cas system. 1. Guide RNA (gRNA) [00233] In some embodiments, the genome editing tool comprises or is used with a guide RNA (gRNA), which can be a dual-guide RNA (dgRNA) or a single-guide RNA (sgRNA). In some embodiments, any one of the first gRNA and/or the at least one additional gRNA comprises a dual-guide RNA (dgRNA) or a single-guide RNA (sgRNA). A guide RNA directs an RNA-guided DNA binding agent to a target sequence. [00234] In some embodiments of the present disclosure, the cargo for the lipid nucleic acid assembly formulation includes at least one gRNA or a nucleic acid encoding the same. The gRNA may guide the Cas nuclease or Class 2 Cas nuclease to a target sequence on a target nucleic acid molecule. In some embodiments, a gRNA binds with and provides specificity of
Attorney Docket: 01155-0064-00PCT cleavage by a Class 2 Cas nuclease. In some embodiments, the gRNA and the Cas nuclease may form a ribonucleoprotein (RNP), e.g., a CRISPR/Cas complex such as a CRISPR/Cas9 complex. In some embodiments, the CRISPR/Cas complex may be a Type-II CRISPR/Cas9 complex. In some embodiments, the CRISPR/Cas complex may be a Type-V CRISPR/Cas complex, such as a Cpf1/guide RNA complex. Cas nucleases and cognate gRNAs may be paired. The gRNA scaffold structures that pair with each Class 2 Cas nuclease vary with the specific CRISPR/Cas system. [00235] In some embodiments, the sgRNA is a “Cas9 sgRNA” capable of mediating RNA- guided DNA cleavage by a Cas9 protein. In some embodiments, the sgRNA is a “Cpf1 sgRNA” capable of mediating RNA-guided DNA cleavage by a Cpf1 protein. In some embodiments, the gRNA comprises a crRNA and tracr RNA sufficient for forming an active complex with a Cas9 protein and mediating RNA-guided DNA cleavage. In some embodiments, the gRNA comprises a crRNA sufficient for forming an active complex with a Cpf1 protein and mediating RNA-guided DNA cleavage. See Zetsche 2015. [00236] Certain embodiments of the disclosure also provide nucleic acids, e.g., expression cassettes, encoding the gRNA described herein. A “guide RNA nucleic acid” is used herein to refer to a guide RNA (e.g. an sgRNA or a dgRNA) and a guide RNA expression cassette, which is a nucleic acid that encodes one or more guide RNAs. [00237] In some embodiments, the nucleic acid may be a DNA molecule. In some embodiments, the nucleic acid may comprise a nucleotide sequence encoding a crRNA. In some embodiments, the nucleotide sequence encoding the crRNA comprises a targeting sequence flanked by all or a portion of a repeat sequence from a naturally-occurring CRISPR/Cas system. In some embodiments, the nucleic acid may comprise a nucleotide sequence encoding a tracr RNA. In some embodiments, the crRNA and the tracr RNA may be encoded by two separate nucleic acids. In other embodiments, the crRNA and the tracr RNA may be encoded by a single nucleic acid. In some embodiments, the crRNA and the tracr RNA may be encoded by opposite strands of a single nucleic acid. In other embodiments, the crRNA and the tracr RNA may be encoded by the same strand of a single nucleic acid. In some embodiments, the gRNA nucleic acid encodes an sgRNA. In some embodiments, the gRNA nucleic acid encodes a Cas9 nuclease sgRNA. In come embodiments, the gRNA nucleic acid encodes a Cpf1 nuclease sgRNA. [00238] The nucleotide sequence encoding the guide RNA may be operably linked to at least one transcriptional or regulatory control sequence, such as a promoter, a 3' UTR, or a 5' UTR. In one example, the promoter may be a tRNA promoter, e.g., tRNA
Lys3, or a tRNA
Attorney Docket: 01155-0064-00PCT chimera. See Mefferd et al., RNA.201521:1683-9; Scherer et al., Nucleic Acids Res.200735: 2620–2628. In some embodiments, the promoter may be recognized by RNA polymerase III (Pol III). Non-limiting examples of Pol III promoters also include U6 and H1 promoters. In some embodiments, the nucleotide sequence encoding the guide RNA may be operably linked to a mouse or human U6 promoter. In some embodiments, the gRNA nucleic acid is a modified nucleic acid. In some embodiments, the gRNA nucleic acid includes a modified nucleoside or nucleotide. In some embodiments, the gRNA nucleic acid includes a 5' end modification, for example a modified nucleoside or nucleotide to stabilize and prevent integration of the nucleic acid. In some embodiments, the gRNA nucleic acid comprises a double-stranded DNA having a 5' end modification on each strand. In some embodiments, the gRNA nucleic acid includes an inverted dideoxy-T or an inverted abasic nucleoside or nucleotide as the 5' end modification. In some embodiments, the gRNA nucleic acid includes a label such as biotin, desthiobiotin- TEG, digoxigenin, and fluorescent markers, including, for example, FAM, ROX, TAMRA, and AlexaFluor. [00239] In some embodiments, more than one gRNA nucleic acid, such as a gRNA, can be used with a CRISPR/Cas nuclease system. Each gRNA nucleic acid may contain a different targeting sequence, such that the CRISPR/Cas system cleaves more than one target sequence. In some embodiments, one or more gRNAs may have the same or differing properties such as activity or stability within a CRISPR/Cas complex. Where more than one gRNA is used, each gRNA can be encoded on the same or on different gRNA nucleic acid. The promoters used to drive expression of the more than one gRNA may be the same or different. [00240] Target sequences for Cas proteins include both the positive and negative strands of genomic DNA (i.e., the sequence given and the sequence’s reverse compliment), as a nucleic acid substrate for a Cas protein is a double stranded nucleic acid. Accordingly, where a guide sequence is said to be “complementary to a target sequence”, it is to be understood that the guide sequence may direct a guide RNA to bind to the reverse complement of a target sequence. Thus, in some embodiments, where the guide sequence binds the reverse complement of a target sequence, the guide sequence is identical to certain nucleotides of the target sequence (e.g., the target sequence not including the PAM) except for the substitution of U for T in the guide sequence. [00241] The length of the targeting sequence may depend on the CRISPR/Cas system and components used. For example, different Class 2 Cas nucleases from different bacterial species have varying optimal targeting sequence lengths. Accordingly, the targeting sequence may comprise 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
Attorney Docket: 01155-0064-00PCT 29, 30, 35, 40, 45, 50, or more than 50 nucleotides in length. In some embodiments, the targeting sequence length is 0, 1, 2, 3, 4, or 5 nucleotides longer or shorter than the guide sequence of a naturally-occurring CRISPR/Cas system. In some embodiments, the Cas nuclease and gRNA scaffold will be derived from the same CRISPR/Cas system. In some embodiments, the targeting sequence may comprise or consist of 18-24 nucleotides. In some embodiments, the targeting sequence may comprise or consist of 19-21 nucleotides. In some embodiments, the targeting sequence may comprise or consist of 20 nucleotides. [00242] In some embodiments, the first gRNA comprises the sequence of SEQ ID NO: 727, a sequence having 90% or 95% identity to SEQ ID NO: 727, or a sequence having at least 17, 18, 19, or 20 contiguous nucleotides of SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 90% or 95% identity to SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 90% identity to SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 95% identity to SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having at least 17, 18, 19, or 20 contiguous nucleotides of SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 17 contiguous nucleotides of SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 18 contiguous nucleotides of SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 19 contiguous nucleotides of SEQ ID NO: 727. In some embodiments, the first gRNA comprises a sequence having 20 contiguous nucleotides of SEQ ID NO: 727. In some embodiments, the first gRNA comprises the sequence of SEQ ID NO: 727. In some embodiments, the first gRNA comprises the sequence of SEQ ID NO: 716. [00243] In some embodiments, the at least one additional gRNA comprises the sequence of SEQ ID NO: 728, a sequence having 90% or 95% identity to SEQ ID NO: 728, or a sequence having at least 17, 18, 19, or 20 contiguous nucleotides of SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having 90% or 95% identity to SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having 90% identity to SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having 95% identity to SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having at least 17, 18, 19, or 20 contiguous nucleotides of SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having 17 contiguous nucleotides of SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having 18 contiguous nucleotides of SEQ ID NO: 728. In some embodiments, the at least one additional
Attorney Docket: 01155-0064-00PCT gRNA comprises a sequence having 19 contiguous nucleotides of SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises a sequence having 20 contiguous nucleotides of SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises the sequence of SEQ ID NO: 728. In some embodiments, the at least one additional gRNA comprises the sequence of SEQ ID NO: 724. 2. RNA-guided DNA binding agent [00244] In some embodiments, the genome editing tool is a RNA-guided DNA binding agent. In some embodiments, the RNA-guided DNA binding agent is a Cas cleavase/nickase and/or an inactivated forms thereof (dCas DNA binding agents). In some embodiments, the RNA-guided DNA binding agent is a Cas nuclease. [00245] In some embodiments, the genome editing tool is a nucleic acid genome editing tool encoding an RNA-guided DNA binding agent. In some embodiments, the genome editing tool is an mRNA encoding an RNA-guided DNA binding agent. In some embodiments, the genome editing tool is an mRNA encoding a Cas nuclease. In some embodiments, the genome editing tool is an mRNA encoding an RNA-guided DNA binding agent and comprising an open reading frame (ORF) comprising any one of SEQ ID NOs: 1-5, 29, and 30 or a sequence having at least 90% identity to any one of SEQ ID NOs: 1-5, 29, and 30. [00246] In some embodiments, genome editing tool comprises a mRNA such as a Cas nuclease mRNA and a gRNA nucleic acid that are co-encapsulated in the lipid nucleic acid assembly composition. In some embodiments, an mRNA encoding a RNA-guided DNA binding agent is formulated in a first lipid nucleic acid assembly composition and a gRNA nucleic acid is formulated in a second lipid nucleic acid assembly composition. In some embodiments, the first and second lipid nucleic acid assembly compositions are administered simultaneously. In other embodiments, the first and second lipid nucleic acid assembly compositions are administered sequentially. In some embodiments, the first and second lipid nucleic acid assembly compositions are combined prior to the preincubation step. In some embodiments, the first and second lipid nucleic acid assembly compositions are preincubated separately. [00247] Non-limiting exemplary species that the Cas nuclease can be derived from include Streptococcus pyogenes, Streptococcus thermophilus, Streptococcus sp., Staphylococcus aureus, Listeria innocua, Lactobacillus gasseri, Francisella novicida, Wolinella succinogenes, Sutterella wadsworthensis, Gammaproteobacterium, Neisseria meningitidis, Campylobacter
Attorney Docket: 01155-0064-00PCT jejuni, Pasteurella multocida, Fibrobacter succinogene, Rhodospirillum rubrum, Nocardiopsis dassonvillei, Streptomyces pristinaespiralis, Streptomyces viridochromogenes, Streptomyces viridochromogenes, Streptosporangium roseum, Streptosporangium roseum, Alicyclobacillus acidocaldarius, Bacillus pseudomycoides, Bacillus selenitireducens, Exiguobacterium sibiricum, Lactobacillus delbrueckii, Lactobacillus salivarius, Lactobacillus buchneri, Treponema denticola, Microscilla marina, Burkholderiales bacterium, Polaromonas naphthalenivorans, Polaromonas sp., Crocosphaera watsonii, Cyanothece sp., Microcystis aeruginosa, Synechococcus sp., Acetohalobium arabaticum, Ammonifex degensii, Caldicelulosiruptor becscii, Candidatus Desulforudis, Clostridium botulinum, Clostridium difficile, Finegoldia magna, Natranaerobius thermophilus, Pelotomaculum thermopropionicum, Acidithiobacillus caldus, Acidithiobacillus ferrooxidans, Allochromatium vinosum, Marinobacter sp., Nitrosococcus halophilus, Nitrosococcus watsoni, Pseudoalteromonas haloplanktis, Ktedonobacter racemifer, Methanohalobium evestigatum, Anabaena variabilis, Nodularia spumigena, Nostoc sp., Arthrospira maxima, Arthrospira platensis, Arthrospira sp., Lyngbya sp., Microcoleus chthonoplastes, Oscillatoria sp., Petrotoga mobilis, Thermosipho africanus, Streptococcus pasteurianus, Neisseria cinerea, Campylobacter lari, Parvibaculum lavamentivorans, Corynebacterium diphtheria, Acidaminococcus sp., Lachnospiraceae bacterium ND2006, and Acaryochloris marina. [00248] In some embodiments, the Cas nuclease is the Cas9 nuclease from Streptococcus pyogenes. In some embodiments, the Cas nuclease is the Cas9 nuclease from Streptococcus thermophilus. In some embodiments, the Cas nuclease is the Cas9 nuclease from Neisseria meningitidis. In some embodiments, the Cas nuclease is the Cas9 nuclease from Staphylococcus aureus. In some embodiments, the Cas nuclease is the Cpf1 nuclease from Francisella novicida. In some embodiments, the Cas nuclease is the Cpf1 nuclease from Acidaminococcus sp. In some embodiments, the Cas nuclease is the Cpf1 nuclease from Lachnospiraceae bacterium ND2006. In further embodiments, the Cas nuclease is the Cpf1 nuclease from Francisella tularensis, Lachnospiraceae bacterium, Butyrivibrio proteoclasticus, Peregrinibacteria bacterium, Parcubacteria bacterium, Smithella, Acidaminococcus, Candidatus Methanoplasma termitum, Eubacterium eligens, Moraxella bovoculi, Leptospira inadai, Porphyromonas crevioricanis, Prevotella disiens, or Porphyromonas macacae. In some embodiments, the Cas nuclease is a Cpf1 nuclease from an Acidaminococcus or Lachnospiraceae. [00249] Wild type Cas9 has two nuclease domains: RuvC and HNH. The RuvC domain cleaves the non-target DNA strand, and the HNH domain cleaves the target strand of DNA. In
Attorney Docket: 01155-0064-00PCT some embodiments, the Cas9 nuclease comprises more than one RuvC domain and/or more than one HNH domain. In some embodiments, the Cas9 nuclease is a wild type Cas9. In some embodiments, the Cas9 is capable of inducing a double strand break in target DNA. In some embodiments, the Cas nuclease may cleave dsDNA, it may cleave one strand of dsDNA, or it may not have DNA cleavase or nickase activity. [00250] In some embodiments, chimeric Cas nucleases are used, where one domain or region of the protein is replaced by a portion of a different protein. In some embodiments, a Cas nuclease domain may be replaced with a domain from a different nuclease such as Fok1. In some embodiments, a Cas nuclease may be a modified nuclease. [00251] In other embodiments, the Cas nuclease or Cas nickase may be from a Type-I CRISPR/Cas system. In some embodiments, the Cas nuclease may be a component of the Cascade complex of a Type-I CRISPR/Cas system. In some embodiments, the Cas nuclease may be a Cas3 protein. In some embodiments, the Cas nuclease may be from a Type-III CRISPR/Cas system. In some embodiments, the Cas nuclease may have an RNA cleavage activity. [00252] In some embodiments, the RNA-guided DNA-binding agent has single-strand nickase activity, i.e., can cut one DNA strand to produce a single-strand break, also known as a “nick.” In some embodiments, the RNA-guided DNA-binding agent comprises a Cas nickase. A nickase is an enzyme that creates a nick in dsDNA, i.e., cuts one strand but not the other of the DNA double helix. In some embodiments, a Cas nickase is a version of a Cas nuclease (e.g., a Cas nuclease discussed above) in which an endonucleolytic active site is inactivated, e.g., by one or more alterations (e.g., point mutations) in a catalytic domain. See, e.g., US Pat. No. 8,889,356 for discussion of Cas nickases and exemplary catalytic domain alterations. In some embodiments, a Cas nickase such as a Cas9 nickase has an inactivated RuvC or HNH domain. [00253] In some embodiments, the RNA-guided DNA-binding agent is modified to contain only one functional nuclease domain. For example, the agent protein may be modified such that one of the nuclease domains is mutated or fully or partially deleted to reduce its nucleic acid cleavage activity. In some embodiments, a nickase is used having a RuvC domain with reduced activity. In some embodiments, a nickase is used having an inactive RuvC domain. In some embodiments, a nickase is used having an HNH domain with reduced activity. In some embodiments, a nickase is used having an inactive HNH domain. [00254] In some embodiments, a conserved amino acid within a Cas protein nuclease domain is substituted to reduce or alter nuclease activity. In some embodiments, a Cas nuclease
Attorney Docket: 01155-0064-00PCT may comprise an amino acid substitution in the RuvC or RuvC-like nuclease domain. Exemplary amino acid substitutions in the RuvC or RuvC-like nuclease domain include D10A (based on the S. pyogenes Cas9 protein). See, e.g., Zetsche et al. (2015) Cell Oct 22:163(3): 759-771. In some embodiments, the Cas nuclease may comprise an amino acid substitution in the HNH or HNH-like nuclease domain. Exemplary amino acid substitutions in the HNH or HNH-like nuclease domain include E762A, H840A, N863A, H983A, and D986A (based on the S. pyogenes Cas9 protein). See, e.g., Zetsche et al. (2015). Further exemplary amino acid substitutions include D917A, E1006A, and D1255A (based on the Francisella novicida U112 Cpf1 (FnCpf1) sequence (UniProtKB - A0Q7Q2 (CPF1_FRATN)). [00255] In some embodiments, an mRNA encoding a nickase is provided in combination with a pair of guide RNAs that are complementary to the sense and antisense strands of the target sequence, respectively. In this embodiment, the guide RNAs direct the nickase to a target sequence and introduce a DSB by generating a nick on opposite strands of the target sequence (i.e., double nicking). In some embodiments, use of double nicking may improve specificity and reduce off-target effects. In some embodiments, a nickase is used together with two separate guide RNAs targeting opposite strands of DNA to produce a double nick in the target DNA. In some embodiments, a nickase is used together with two separate guide RNAs that are selected to be in close proximity to produce a double nick in the target DNA. [00256] In some embodiments, the RNA-guided DNA-binding agent lacks cleavase and nickase activity. In some embodiments, the RNA-guided DNA-binding agent comprises a dCas DNA-binding polypeptide. A dCas polypeptide has DNA-binding activity while essentially lacking catalytic (cleavase/nickase) activity. In some embodiments, the dCas polypeptide is a dCas9 polypeptide. In some embodiments, the RNA-guided DNA-binding agent lacking cleavase and nickase activity or the dCas DNA-binding polypeptide is a version of a Cas nuclease (e.g., a Cas nuclease discussed above) in which its endonucleolytic active sites are inactivated, e.g., by one or more alterations (e.g., point mutations) in its catalytic domains. See, e.g., US 2014/0186958 A1; US 2015/0166980 A1. [00257] In some embodiments, the RNA-guided DNA binding agent comprises a APOBEC3 deaminase. In some embodiments, a APOBEC3 deaminase is a APOBEC3A (A3A). In some embodiments, the A3A is a human A3A. In some embodiments, the A3A is a wild-type A3A. [00258] In some embodiments, the RNA-guided DNA binding agent comprises an editor. An exemplary editor is BC22n which comprises a H. sapiens APOBEC3A fused to S. pyogenes-D10A Cas9 nickase by an XTEN linker.
Attorney Docket: 01155-0064-00PCT [00259] In some embodiments, the RNA-guided DNA-binding agent comprises one or more heterologous functional domains (e.g., is or comprises a fusion polypeptide). [00260] In some embodiments, the heterologous functional domain may facilitate transport of the RNA-guided DNA-binding agent into the nucleus of a cell. For example, the heterologous functional domain may be a nuclear localization signal (NLS). In some embodiments, the RNA-guided DNA-binding agent may be fused with 1-10 NLS(s). In some embodiments, the RNA-guided DNA-binding agent may be fused with 1-5 NLS(s). In some embodiments, the RNA-guided DNA-binding agent may be fused with one NLS. Where one NLS is used, the NLS may be fused at the N-terminus or the C-terminus of the RNA-guided DNA-binding agent sequence. It may also be inserted within the RNA-guided DNA binding agent sequence. In other embodiments, the RNA-guided DNA-binding agent may be fused with more than one NLS. In some embodiments, the RNA-guided DNA-binding agent may be fused with 2, 3, 4, or 5 NLSs. In some embodiments, the RNA-guided DNA-binding agent may be fused with two NLSs. In some circumstances, the two NLSs may be the same (e.g., two SV40 NLSs) or different. In some embodiments, the RNA-guided DNA-binding agent is fused to two NLS sequences (e.g., SV40) fused at the carboxy terminus. In some embodiments, the RNA-guided DNA-binding agent may be fused with two NLSs, one linked at the N-terminus and one at the C-terminus. In some embodiments, the RNA-guided DNA-binding agent may be fused with 3 NLSs. In some embodiments, the RNA-guided DNA-binding agent may be fused with no NLS. In some embodiments, the NLS may be a monopartite sequence, such as, e.g., the SV40 NLS, PKKKRKV (SEQ ID NO: 23) or PKKKRRV (SEQ ID NO: 24). In some embodiments, the NLS may be a bipartite sequence, such as the NLS of nucleoplasmin, KRPAATKKAGQAKKKK (SEQ ID NO: 25). In a specific embodiment, a single PKKKRKV (SEQ ID NO: 23) NLS may be fused at the C-terminus of the RNA-guided DNA-binding agent. One or more linkers are optionally included at the fusion site. [00261] In some embodiments, the heterologous functional domain may be capable of modifying the intracellular half-life of the RNA-guided DNA binding agent. In some embodiments, the half-life of the RNA-guided DNA binding agent may be increased. In some embodiments, the half-life of the RNA-guided DNA-binding agent may be reduced. In some embodiments, the heterologous functional domain may be capable of increasing the stability of the RNA-guided DNA-binding agent. In some embodiments, the heterologous functional domain may be capable of reducing the stability of the RNA-guided DNA-binding agent. In some embodiments, the heterologous functional domain may act as a signal peptide for protein degradation. In some embodiments, the protein degradation may be mediated by proteolytic
Attorney Docket: 01155-0064-00PCT enzymes, such as, for example, proteasomes, lysosomal proteases, or calpain proteases. In some embodiments, the heterologous functional domain may comprise a PEST sequence. In some embodiments, the RNA-guided DNA-binding agent may be modified by addition of ubiquitin or a polyubiquitin chain. In some embodiments, the ubiquitin may be a ubiquitin- like protein (UBL). Non-limiting examples of ubiquitin-like proteins include small ubiquitin- like modifier (SUMO), ubiquitin cross-reactive protein (UCRP, also known as interferon- stimulated gene-15 (ISG15)), ubiquitin-related modifier-1 (URM1), neuronal-precursor-cell- expressed developmentally downregulated protein-8 (NEDD8, also called Rub1 in S. cerevisiae), human leukocyte antigen F-associated (FAT10), autophagy-8 (ATG8) and -12 (ATG12), Fau ubiquitin-like protein (FUB1), membrane-anchored UBL (MUB), ubiquitin fold-modifier-1 (UFM1), and ubiquitin-like protein-5 (UBL5). [00262] In some embodiments, the heterologous functional domain may be a marker domain. Non-limiting examples of marker domains include fluorescent proteins, purification tags, epitope tags, and reporter gene sequences. In some embodiments, the marker domain may be a fluorescent protein. Non-limiting examples of suitable fluorescent proteins include green fluorescent proteins (e.g., GFP, GFP-2, tagGFP, turboGFP, sfGFP, EGFP, Emerald, Azami Green, Monomeric Azami Green, CopGFP, AceGFP, ZsGreen1 ), yellow fluorescent proteins (e.g., YFP, EYFP, Citrine, Venus, YPet, PhiYFP, ZsYellow1), blue fluorescent proteins (e.g., EBFP, EBFP2, Azurite, mKalamal, GFPuv, Sapphire, T-sapphire,), cyan fluorescent proteins (e.g., ECFP, Cerulean, CyPet, AmCyan1, Midoriishi-Cyan), red fluorescent proteins (e.g., mKate, mKate2, mPlum, DsRed monomer, mCherry, mRFP1, DsRed-Express, DsRed2, DsRed-Monomer, HcRed-Tandem, HcRed1, AsRed2, eqFP611, mRasberry, mStrawberry, Jred), and orange fluorescent proteins (mOrange, mKO, Kusabira- Orange, Monomeric Kusabira-Orange, mTangerine, tdTomato) or any other suitable fluorescent protein. In other embodiments, the marker domain may be a purification tag and/or an epitope tag. Non-limiting exemplary tags include glutathione-S-transferase (GST), chitin binding protein (CBP), maltose binding protein (MBP), thioredoxin (TRX), poly(NANP), tandem affinity purification (TAP) tag, myc, AcV5, AU1, AU5, E, ECS, E2, FLAG, HA, nus, Softag 1, Softag 3, Strep, SBP, Glu-Glu, HSV, KT3, S, S1, T7, V5, VSV-G, 6xHis, 8xHis, biotin carboxyl carrier protein (BCCP), poly-His, and calmodulin. Non-limiting exemplary reporter genes include glutathione-S-transferase (GST), horseradish peroxidase (HRP), chloramphenicol acetyltransferase (CAT), beta-galactosidase, beta-glucuronidase, luciferase, or fluorescent proteins.
Attorney Docket: 01155-0064-00PCT [00263] In additional embodiments, the heterologous functional domain may target the RNA-guided DNA-binding agent to a specific organelle, cell type, tissue, or organ. In some embodiments, the heterologous functional domain may target the RNA-guided DNA-binding agent to mitochondria. [00264] In further embodiments, the heterologous functional domain may be an effector domain such as an editor domain. When the RNA-guided DNA-binding agent is directed to its target sequence, e.g., when a Cas nuclease is directed to a target sequence by a gRNA, the effector domain such as an editor domain may modify or affect the target sequence. In some embodiments, the effector domain such as an editor domain may be chosen from a nucleic acid binding domain, a nuclease domain (e.g., a non-Cas nuclease domain), an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. In some embodiments, the heterologous functional domain is a nuclease, such as a FokI nuclease. See, e.g., US Pat. No. 9,023,649. In some embodiments, the heterologous functional domain is a transcriptional activator or repressor. See, e.g., Qi et al., “Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression,” Cell 152:1173-83 (2013); Perez-Pinera et al., “RNA-guided gene activation by CRISPR-Cas9- based transcription factors,” Nat. Methods 10:973-6 (2013); Mali et al., “CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering,” Nat. Biotechnol. 31:833-8 (2013); Gilbert et al., “CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes,” Cell 154:442-51 (2013). As such, the RNA-guided DNA-binding agent essentially becomes a transcription factor that can be directed to bind a desired target sequence using a guide RNA. In some embodiments, the DNA modification domain is a methylation domain, such as a demethylation or methyltransferase domain. In some embodiments, the effector domain is a DNA modification domain, such as a base-editing domain. In particular embodiments, the DNA modification domain is a nucleic acid editing domain that introduces a specific modification into the DNA, such as a deaminase domain. See, e.g., WO 2015/089406; US 2016/0304846. The nucleic acid editing domains, deaminase domains, and Cas9 variants described in WO 2015/089406 and U.S.2016/0304846 are hereby incorporated by reference. [00265] The nuclease may comprise at least one domain that interacts with a guide RNA (“gRNA”). Additionally, the nuclease may be directed to a target sequence by a gRNA. In Class 2 Cas nuclease systems, the gRNA interacts with the nuclease as well as the target sequence, such that it directs binding to the target sequence. In some embodiments, the gRNA provides the specificity for the targeted cleavage, and the nuclease may be universal and paired
Attorney Docket: 01155-0064-00PCT with different gRNAs to cleave different target sequences. Class 2 Cas nuclease may pair with a gRNA scaffold structure of the types, orthologs, and exemplary species listed above. B. Additional Genome Editing System Tools [00266] In some embodiments, the genome editing tool is a component of a genome editing system chosen from a zinc finger nuclease system, a TALEN system, and a meganuclease system. In some embodiments, the genome editing tool is a nucleic acid encoding one or more components of such genome editing system. Exemplary components of the system include meganucleases, zinc finger nucleases, TALENS, and fragments thereof. [00267] In some embodiments, the gene editing system is a TALEN system. Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA strands). Transcription activator-like effectors (TALEs) can be engineered to bind to a desired DNA sequence, to promote DNA cleavage at specific locations (see, e.g., Boch, 2011, Nature Biotech). The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing in situ, a technique known as genome editing with engineered nucleases. Such methods and compositions for use therein are known in the art. See, e.g., WO2019147805, WO2014040370, WO2018073393, the contents of which are hereby incorporated in their entireties. [00268] In some embodiments, the gene editing system is a zinc-finger system. Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA- binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences to enables zinc-finger nucleases to target unique sequences within complex genomes. The non-specific cleavage domain from the type IIs restriction endonuclease FokI is typically used as the cleavage domain in ZFNs. Cleavage is repaired by endogenous DNA repair machinery, allowing ZFN to precisely alter the genomes of higher organisms. Such methods and compositions for use therein are known in the art. See, e.g., WO2011091324, the contents of which are hereby incorporated in their entireties. V. Exemplary Nucleic Acids for Lipid Nucleic Acid Assembly Compositions [00269] In some embodiments, the lipid nucleic acid assembly compositions (e.g., lipid nanoparticles (“LNPs”)) deliver a nucleic acid (or polynucleotide) to a cell. In some embodiments, the nucleic acid comprises nucleosides or nucleoside analogs which have
Attorney Docket: 01155-0064-00PCT nitrogenous heterocyclic bases or base analogs linked together along a backbone, including conventional RNA, DNA, mixed RNA-DNA, and polymers that are analogs thereof. A. Modified Nucleic Acids [00270] In some embodiments, the lipid nucleic acid assembly compositions comprise modified RNAs. In some embodiments, the lipid nucleic acid assembly compositions comprise modified DNAs. [00271] Modified nucleosides or nucleotides can be present in an RNA, for example a gRNA or mRNA. A gRNA or mRNA comprising one or more modified nucleosides or nucleotides, for example, is called a “modified” RNA to describe the presence of one or more non-naturally and/or naturally occurring components or configurations that are used instead of or in addition to the canonical A, G, C, and U residues. In some embodiments, a modified RNA is synthesized with a non-canonical nucleoside or nucleotide, here called “modified.” [00272] Modified nucleosides and nucleotides can include one or more of: (i) alteration, e.g., replacement, of one or both of the non-linking phosphate oxygens and/or of one or more of the linking phosphate oxygens in the phosphodiester backbone linkage (an exemplary backbone modification); (ii) alteration, e.g., replacement, of a constituent of the ribose sugar, e.g., of the 2' hydroxyl on the ribose sugar (an exemplary sugar modification); (iii) wholesale replacement of the phosphate moiety with “dephospho” linkers (an exemplary backbone modification); (iv) modification or replacement of a naturally occurring nucleobase, including with a non-canonical nucleobase (an exemplary base modification); (v) replacement or modification of the ribose-phosphate backbone (an exemplary backbone modification); (vi) modification of the 3' end or 5' end of the oligonucleotide, e.g., removal, modification or replacement of a terminal phosphate group or conjugation of a moiety, cap or linker (such 3' or 5' cap modifications may comprise a sugar and/or backbone modification); and (vii) modification or replacement of the sugar (an exemplary sugar modification). Certain embodiments comprise a 5' end modification to an mRNA, gRNA, or nucleic acid. Certain embodiments comprise a 3' end modification to an mRNA, gRNA, or nucleic acid. A modified RNA can contain 5' end and 3' end modifications. A modified RNA can contain one or more modified residues at non-terminal locations. In some embodiments, a gRNA includes at least one modified residue. In some embodiments, an mRNA includes at least one modified residue. [00273] As used herein, a first sequence is considered to “comprise a sequence with at least X% identity to” a second sequence if an alignment of the first sequence to the second sequence shows that X% or more of the positions of the second sequence in its entirety are matched by
Attorney Docket: 01155-0064-00PCT the first sequence. For example, the sequence AAGA comprises a sequence with 100% identity to the sequence AAG because an alignment would give 100% identity in that there are matches to all three positions of the second sequence. The differences between RNA and DNA (generally the exchange of uridine for thymidine or vice versa) and the presence of nucleoside analogs such as modified uridines do not contribute to differences in identity or complementarity among polynucleotides as long as the relevant nucleotides (such as thymidine, uridine, or modified uridine) have the same complement (e.g., adenosine for all of thymidine, uridine, or modified uridine; another example is cytosine and 5-methylcytosine, both of which have guanosine or modified guanosine as a complement). Thus, for example, the sequence 5’-AXG where X is any modified uridine, such as pseudouridine, N1-methyl pseudouridine, or 5-methoxyuridine, is considered 100% identical to AUG in that both are perfectly complementary to the same sequence (5’-CAU). Exemplary alignment algorithms are the Smith-Waterman and Needleman-Wunsch algorithms, which are well-known in the art. One skilled in the art will understand what choice of algorithm and parameter settings are appropriate for a given pair of sequences to be aligned; for sequences of generally similar length and expected identity >50% for amino acids or >75% for nucleotides, the Needleman- Wunsch algorithm with default settings of the Needleman-Wunsch algorithm interface provided by the EBI at the www.ebi.ac.uk web server is generally appropriate. [00274] In some embodiments, a composition or formulation disclosed herein comprises an mRNA comprising an open reading frame (ORF), such as, e.g. an ORF encoding an RNA- guided DNA binding agent, such as a Cas nuclease, or Class 2 Cas nuclease as described herein. In some embodiments, an mRNA comprising an ORF encoding an RNA-guided DNA binding agent, such as a Cas nuclease or Class 2 Cas nuclease, is provided, used, or administered. In some embodiments, the ORF is codon optimized. In some embodiments, the ORF encoding an RNA-guided DNA binding agent is a “modified RNA-guided DNA binding agent ORF” or simply a “modified ORF,” which is used as shorthand to indicate that the ORF is modified in one or more of the following ways: (1) the modified ORF has a uridine content ranging from its minimum uridine content to 150% of the minimum uridine content; (2) the modified ORF has a uridine dinucleotide content ranging from its minimum uridine dinucleotide content to 150% of the minimum uridine dinucleotide content; (3) the modified ORF has at least 90% identity to any one of any of the Cas ORFs in Table 6; (4) the modified ORF consists of a set of codons of which at least 75% of the codons are minimal uridine codon(s) for a given amino acid, e.g. the codon(s) with the fewest uridines (usually 0 or 1 except for a codon for phenylalanine, where the minimal uridine codon has 2 uridines); or (5) the modified ORF
Attorney Docket: 01155-0064-00PCT comprises at least one modified uridine. In some embodiments, the modified ORF is modified in at least two, three, or four of the foregoing ways. In some embodiments, the modified ORF comprises at least one modified uridine and is modified in at least one, two, three, or all of (1)- (4) above. [00275] “Modified uridine” is used herein to refer to a nucleoside other than thymidine with the same hydrogen bond acceptors as uridine and one or more structural differences from uridine. In some embodiments, a modified uridine is a substituted uridine, i.e., a uridine in which one or more non-proton substituents (e.g., alkoxy, such as methoxy) takes the place of a proton. In some embodiments, a modified uridine is pseudouridine. In some embodiments, a modified uridine is a substituted pseudouridine, i.e., a pseudouridine in which one or more non- proton substituents (e.g., alkyl, such as methyl) takes the place of a proton. In some embodiments, a modified uridine is any of a substituted uridine, pseudouridine, or a substituted pseudouridine. [00276] “Uridine position” as used herein refers to a position in a polynucleotide occupied by a uridine or a modified uridine. Thus, for example, a polynucleotide in which “100% of the uridine positions are modified uridines” contains a modified uridine at every position that would be a uridine in a conventional RNA (where all bases are standard A, U, C, or G bases) of the same sequence. Unless otherwise indicated, a U in a polynucleotide sequence of a sequence table or sequence listing in, or accompanying, this disclosure can be a uridine or a modified uridine. [00277] Minimal uridine codons: Amino Acid Minimal uridine codon A Al i GCA GCC GCG
Attorney Docket: 01155-0064-00PCT F Phenylalanine UUC Y Tyrosine UAC [00278] In a

consist of a set of codons of which at least 75%, 80%, 85%, 90%, 95%, 98%, 99%, or 100% of the codons are codons listed in the Table above of minimal uridine codons. In any of the foregoing embodiments, the modified ORF may comprise a sequence with at least 90%, 95%, 98%, 99%, or 100% identity to any one of the Cas ORFs in Table 6. [00279] In any of the foregoing embodiments, the modified ORF may have a uridine content ranging from its minimum uridine content to 150%, 145%, 140%, 135%, 130%, 125%, 120%, 115%, 110%, 105%, 104%, 103%, 102%, or 101% of the minimum uridine content. [00280] In any of the foregoing embodiments, the modified ORF may have a uridine dinucleotide content ranging from its minimum uridine dinucleotide content to 150%, 145%, 140%, 135%, 130%, 125%, 120%, 115%, 110%, 105%, 104%, 103%, 102%, or 101% of the minimum uridine dinucleotide content. [00281] In any of the foregoing embodiments, the modified ORF may comprise a modified uridine at least at one, a plurality of, or all uridine positions. In some embodiments, the modified uridine is a uridine modified at the 5 position, e.g., with a halogen, methyl, or ethyl. In some embodiments, the modified uridine is a pseudouridine modified at the 1 position, e.g., with a halogen, methyl, or ethyl. The modified uridine can be, for example, pseudouridine, N1- methyl-pseudouridine, 5-methoxyuridine, 5-iodouridine, or a combination thereof. In some embodiments, the modified uridine is 5-methoxyuridine. In some embodiments, the modified uridine is 5-iodouridine. In some embodiments, the modified uridine is pseudouridine. In some embodiments, the modified uridine is N1-methyl-pseudouridine. In some embodiments, the modified uridine is a combination of pseudouridine and N1-methyl-pseudouridine. In some embodiments, the modified uridine is a combination of pseudouridine and 5-methoxyuridine. In some embodiments, the modified uridine is a combination of N1-methyl pseudouridine and 5-methoxyuridine. In some embodiments, the modified uridine is a combination of 5- iodouridine and N1-methyl-pseudouridine. In some embodiments, the modified uridine is a combination of pseudouridine and 5-iodouridine. In some embodiments, the modified uridine is a combination of 5-iodouridine and 5-methoxyuridine.
Attorney Docket: 01155-0064-00PCT [00282] In some embodiments, at least 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 98%, 99%, or 100% of the uridine positions in an mRNA according to the disclosure are modified uridines. In some embodiments, 10%- 25%, 15-25%, 25-35%, 35-45%, 45-55%, 55-65%, 65-75%, 75-85%, 85-95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are modified uridines, e.g., 5- methoxyuridine, 5-iodouridine, N1-methyl pseudouridine, pseudouridine, or a combination thereof. In some embodiments, 10%-25%, 15-25%, 25-35%, 35-45%, 45-55%, 55-65%, 65- 75%, 75-85%, 85-95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are 5-methoxyuridine. In some embodiments, 10%-25%, 15-25%, 25-35%, 35-45%, 45-55%, 55-65%, 65-75%, 75-85%, 85-95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are pseudouridine. In some embodiments, 10%-25%, 15-25%, 25- 35%, 35-45%, 45-55%, 55-65%, 65-75%, 75-85%, 85-95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are N1-methyl pseudouridine. In some embodiments, 10%-25%, 15-25%, 25-35%, 35-45%, 45-55%, 55-65%, 65-75%, 75-85%, 85- 95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are 5- iodouridine. In some embodiments, 10%-25%, 15-25%, 25-35%, 35-45%, 45-55%, 55-65%, 65-75%, 75-85%, 85-95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are 5-methoxyuridine, and the remainder are N1-methyl pseudouridine. In some embodiments, 10%-25%, 15-25%, 25-35%, 35-45%, 45-55%, 55-65%, 65-75%, 75-85%, 85- 95%, or 90-100% of the uridine positions in an mRNA according to the disclosure are 5- iodouridine, and the remainder are N1-methyl pseudouridine. [00283] In any of the foregoing embodiments, the modified ORF may comprise a reduced uridine dinucleotide content, such as the lowest possible uridine dinucleotide (UU) content , e.g. an ORF that (a) uses a minimal uridine codon (as discussed above) at every position and (b) encodes the same amino acid sequence as the given ORF. The uridine dinucleotide (UU) content can be expressed in absolute terms as the enumeration of UU dinucleotides in an ORF or on a rate basis as the percentage of positions occupied by the uridines of uridine dinucleotides (for example, AUUAU would have a uridine dinucleotide content of 40% because 2 of 5 positions are occupied by the uridines of a uridine dinucleotide). Modified uridine residues are considered equivalent to uridines for the purpose of evaluating minimum uridine dinucleotide content. [00284] In some embodiments, the mRNA comprises at least one UTR from an expressed mammalian mRNA, such as a constitutively expressed mRNA. An mRNA is considered constitutively expressed in a mammal if it is continually transcribed in at least one tissue of a
Attorney Docket: 01155-0064-00PCT healthy adult mammal. In some embodiments, the mRNA comprises a 5’ UTR, 3’ UTR, or 5’ and 3’ UTRs from an expressed mammalian RNA, such as a constitutively expressed mammalian mRNA. Actin mRNA is an example of a constitutively expressed mRNA. [00285] In some embodiments, the mRNA comprises at least one UTR from Hydroxysteroid 17-Beta Dehydrogenase 4 (HSD17B4 or HSD), e.g., a 5’ UTR from HSD. In some embodiments, the mRNA comprises at least one UTR from a globin mRNA, for example, human alpha globin (HBA) mRNA, human beta globin (HBB) mRNA, or Xenopus laevis beta globin (XBG) mRNA. In some embodiments, the mRNA comprises a 5’ UTR, 3’ UTR, or 5’ and 3’ UTRs from a globin mRNA, such as HBA, HBB, or XBG. In some embodiments, the mRNA comprises a 5’ UTR from bovine growth hormone, cytomegalovirus (CMV), mouse Hba-a1, HSD, an albumin gene, HBA, HBB, or XBG. In some embodiments, the mRNA comprises a 3’ UTR from bovine growth hormone, cytomegalovirus, mouse Hba-a1, HSD, an albumin gene, HBA, HBB, or XBG. In some embodiments, the mRNA comprises 5’ and 3’ UTRs from bovine growth hormone, cytomegalovirus, mouse Hba-a1, HSD, an albumin gene, HBA, HBB, XBG, heat shock protein 90 (Hsp90), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), beta-actin, alpha-tubulin, tumor protein (p53), or epidermal growth factor receptor (EGFR). [00286] In some embodiments, the mRNA comprises 5’ and 3’ UTRs that are from the same source, e.g., a constitutively expressed mRNA such as actin, albumin, or a globin such as HBA, HBB, or XBG. [00287] In some embodiments, the mRNA does not comprise a 5’ UTR, e.g., there are no additional nucleotides between the 5’ cap and the start codon. In some embodiments, the mRNA comprises a Kozak sequence (described below) between the 5’ cap and the start codon, but does not have any additional 5’ UTR. In some embodiments, the mRNA does not comprise a 3’ UTR, e.g., there are no additional nucleotides between the stop codon and the poly-A tail. [00288] In some embodiments, the mRNA comprises a Kozak sequence. The Kozak sequence can affect translation initiation and the overall yield of a polypeptide translated from an mRNA. A Kozak sequence includes a methionine codon that can function as the start codon. A minimal Kozak sequence is NNNRUGN wherein at least one of the following is true: the first N is A or G and the second N is G. In the context of a nucleotide sequence, R means a purine (A or G). In some embodiments, the Kozak sequence is RNNRUGN, NNNRUGG, RNNRUGG, RNNAUGN, NNNAUGG, or RNNAUGG. In some embodiments, the Kozak sequence is rccRUGg with zero mismatches or with up to one or two mismatches to positions in lowercase. In some embodiments, the Kozak sequence is rccAUGg with zero mismatches or
Attorney Docket: 01155-0064-00PCT with up to one or two mismatches to positions in lowercase. In some embodiments, the Kozak sequence is gccRccAUGG (SEQ ID NO: 26) with zero mismatches or with up to one, two, or three mismatches to positions in lowercase. In some embodiments, the Kozak sequence is gccAccAUG with zero mismatches or with up to one, two, three, or four mismatches to positions in lowercase. In some embodiments, the Kozak sequence is GCCACCAUG. In some embodiments, the Kozak sequence is gccgccRccAUGG (SEQ ID NO: 27) with zero mismatches or with up to one, two, three, or four mismatches to positions in lowercase. [00289] In some embodiments, the mRNA comprising an ORF encoding an RNA-guided DNA binding agent comprises a sequence having at least 90% identity to any of the Cas ORFs in Table 6. [00290] In some embodiments, an mRNA disclosed herein comprises a 5’ cap, such as a Cap0, Cap1, or Cap2. A 5’ cap is generally a 7-methylguanine ribonucleotide (which may be further modified, as discussed below e.g. with respect to ARCA) linked through a 5’- triphosphate to the 5’ position of the first nucleotide of the 5’-to-3’ chain of the mRNA, i.e., the first cap-proximal nucleotide. In Cap0, the riboses of the first and second cap-proximal nucleotides of the mRNA both comprise a 2’-hydroxyl. In Cap1, the riboses of the first and second transcribed nucleotides of the mRNA comprise a 2’-methoxy and a 2’-hydroxyl, respectively. In Cap2, the riboses of the first and second cap-proximal nucleotides of the mRNA both comprise a 2’-methoxy. See, e.g., Katibah et al. (2014) Proc Natl Acad Sci USA 111(33):12025-30; Abbas et al. (2017) Proc Natl Acad Sci USA 114(11):E2106-E2115. Most endogenous higher eukaryotic mRNAs, including mammalian mRNAs such as human mRNAs, comprise Cap1 or Cap2. Cap0 and other cap structures differing from Cap1 and Cap2 may be immunogenic in mammals, such as humans, due to recognition as “non-self” by components of the innate immune system such as IFIT-1 and IFIT-5, which can result in elevated cytokine levels including type I interferon. Components of the innate immune system such as IFIT-1 and IFIT-5 may also compete with eIF4E for binding of an mRNA with a cap other than Cap1 or Cap2, potentially inhibiting translation of the mRNA. [00291] A cap can be included co-transcriptionally. For example, ARCA (anti-reverse cap analog; Thermo Fisher Scientific Cat. No. AM8045) is a cap analog comprising a 7- methylguanine 3’-methoxy-5’-triphosphate linked to the 5’ position of a guanine ribonucleotide which can be incorporated in vitro into a transcript at initiation. ARCA results in a Cap0 cap in which the 2’ position of the first cap-proximal nucleotide is hydroxyl. See, e.g., Stepinski et al., (2001) “Synthesis and properties of mRNAs containing the novel ‘anti-
Attorney Docket: 01155-0064-00PCT reverse’ cap analogs 7-methyl(3'-O-methyl)GpppG and 7-methyl(3'deoxy)GpppG,” RNA 7: 1486–1495. The ARCA structure is shown below. TriLink Biotechnologies Cat. No.
or ppp TriLink Biotechnologies Cat. No. N-7133) can be used to provide a Cap1 structure co-transcriptionally.3’-O-methylated versions of CleanCap
TM AG and CleanCap
TM GG are also available from TriLink Biotechnologies as Cat. Nos. N-7413 and N-7433, respectively. The CleanCap
TM AG structure is shown below.

transcriptionally. For example, Vaccinia capping enzyme is commercially available (New England Biolabs Cat. No. M2080S) and has RNA triphosphatase and guanylyltransferase activities, provided by its D1 subunit, and guanine methyltransferase, provided by its D12 subunit. As such, it can add a 7- methylguanine to an RNA, so as to give Cap0, in the presence of S-adenosyl methionine and GTP. See, e.g., Guo, P. and Moss, B. (1990) Proc. Natl. Acad. Sci. USA 87, 4023-4027; Mao, X. and Shuman, S. (1994) J. Biol. Chem.269, 24472-24479. [00294] In some embodiments, the mRNA further comprises a poly-adenylated (poly-A) tail. In some embodiments, the poly-A tail comprises at least 20, 30, 40, 50, 60, 70, 80, 90, or 100 adenines, optionally up to 300 adenines. In some embodiments, the poly-A tail comprises 95, 96, 97, 98, 99, or 100 adenine nucleotides. In some instances, the poly-A tail is “interrupted” with one or more non-adenine nucleotide “anchors” at one or more locations within the poly-A tail. The poly-A tails may comprise at least 8 consecutive adenine nucleotides, but also comprise one or more non-adenine nucleotide. As used herein, “non-
Attorney Docket: 01155-0064-00PCT adenine nucleotides” refer to any natural or non-natural nucleotides that do not comprise adenine. Guanine, thymine, and cytosine nucleotides are exemplary non-adenine nucleotides. Thus, the poly-A tails on the mRNA described herein may comprise consecutive adenine nucleotides located 3’ to nucleotides encoding an RNA-guided DNA-binding agent or a sequence of interest. In some instances, the poly-A tails on mRNA comprise non-consecutive adenine nucleotides located 3’ to nucleotides encoding an RNA-guided DNA-binding agent or a sequence of interest, wherein non-adenine nucleotides interrupt the adenine nucleotides at regular or irregularly spaced intervals. [00295] As used herein, “non-adenine nucleotides” refer to any natural or non-natural nucleotides that do not comprise adenine. Guanine, thymine, and cytosine nucleotides are exemplary non-adenine nucleotides. Thus, the poly-A tails on the mRNA described herein may comprise consecutive adenine nucleotides located 3’ to nucleotides encoding an RNA-guided DNA-binding agent or a sequence of interest. In some instances, the poly-A tails on mRNA comprise non-consecutive adenine nucleotides located 3’ to nucleotides encoding an RNA- guided DNA-binding agent or a sequence of interest, wherein non-adenine nucleotides interrupt the adenine nucleotides at regular or irregularly spaced intervals. [00296] In some embodiments, the mRNA is purified. In some embodiments, the mRNA is purified using a precipitation method (e.g., LiCl precipitation, alcohol precipitation, or an equivalent method, e.g., as described herein). In some embodiments, the mRNA is purified using a chromatography-based method, such as an HPLC-based method or an equivalent method (e.g., as described herein). In some embodiments, the mRNA is purified using both a precipitation method (e.g., LiCl precipitation) and an HPLC-based method. [00297] In some embodiments, at least one gRNA is provided in combination with an mRNA disclosed herein. In some embodiments, a gRNA is provided as a separate molecule from the mRNA. In some embodiments, a gRNA is provided as a part, such as a part of a UTR, of an mRNA disclosed herein. B. Chemically Modified Nucleic Acids [00298] In some embodiments, the nucleic acid is an RNA, such as a chemically modified RNA. In some embodiments, the nucleic acid is a DNA, or comprises DNA, such as a chemically modified DNA. [00299] An RNA comprising one or more modified nucleosides or nucleotides is called a “modified” RNA or “chemically modified” RNA, to describe the presence of one or more non- naturally and/or naturally occurring components or configurations that are used instead of or
Attorney Docket: 01155-0064-00PCT in addition to the canonical A, G, C, and U residues. In some embodiments, a modified RNA is synthesized with a non-canonical nucleoside or nucleotide, is here called “modified.” Modified nucleosides and nucleotides can include one or more of: (i) alteration, e.g., replacement, of one or both of the non-linking phosphate oxygens and/or of one or more of the linking phosphate oxygens in the phosphodiester backbone linkage (an exemplary backbone modification); (ii) alteration, e.g., replacement, of a constituent of the ribose sugar, e.g., of the 2' hydroxyl on the ribose sugar (an exemplary sugar modification); (iii) wholesale replacement of the phosphate moiety with “dephospho” linkers (an exemplary backbone modification); (iv) modification or replacement of a naturally occurring nucleobase, including with a non- canonical nucleobase (an exemplary base modification); (v) replacement or modification of the ribose-phosphate backbone (an exemplary backbone modification); (vi) modification of the 3' end or 5' end of the oligonucleotide, e.g., removal, modification or replacement of a terminal phosphate group or conjugation of a moiety, cap or linker (such 3' or 5' cap modifications may comprise a sugar and/or backbone modification); and (vii) modification or replacement of the sugar (an exemplary sugar modification). [00300] A gRNA comprising one or more modified nucleosides or nucleotides is called a “modified” gRNA or “chemically modified” RNA, to describe the presence of one or more non-naturally and/or naturally occurring components or configurations that are used instead of or in addition to the canonical A, G, C, and U residues. In some embodiments, a modified gRNA is synthesized with a non-canonical nucleoside or nucleotide, is here called “modified.” [00301] Chemical modifications such as those listed above can be combined to provide modified nucleic acids, DNAs, RNAs, or gRNAs comprising nucleosides and nucleotides (collectively “residues”) that can have two, three, four, or more modifications. For example, a modified residue can have a modified sugar and a modified nucleobase. In some embodiments, every base of a gRNA is modified, e.g., all bases have a modified phosphate group, such as a phosphorothioate group. In some embodiments, all, or substantially all, of the phosphate groups of a gRNA molecule are replaced with phosphorothioate groups. In some embodiments, modified gRNAs comprise at least one modified residue at or near the 5' end of the RNA. In some embodiments, modified gRNAs comprise at least one modified residue at or near the 3' end of the RNA. [00302] In some embodiments, the nucleic acid such as a gRNA comprises one, two, three or more modified residues. In some embodiments, at least 5% (e.g., at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%,
Attorney Docket: 01155-0064-00PCT at least 85%, at least 90%, at least 95%, or 100%) of the positions in a modified gRNA are modified nucleosides or nucleotides. [00303] Unmodified nucleic acids can be prone to degradation by, e.g., intracellular nucleases or those found in serum. For example, nucleases can hydrolyze nucleic acid phosphodiester bonds. Accordingly, in one aspect the modified nucleic acids such as the gRNAs described herein can contain one or more modified nucleosides or nucleotides, e.g., to introduce stability toward intracellular or serum-based nucleases. In some embodiments, the modified gRNA molecules described herein can exhibit a reduced innate immune response when introduced into a population of cells, both in vivo and ex vivo. The term “innate immune response” includes a cellular response to exogenous nucleic acids, including single stranded nucleic acids, which involves the induction of cytokine expression and release, particularly the interferons, and cell death. [00304] In some embodiments of a backbone modification, the phosphate group of a modified residue can be modified by replacing one or more of the oxygens with a different substituent. Further, the modified residue, e.g., modified residue present in a modified nucleic acid, can include the wholesale replacement of an unmodified phosphate moiety with a modified phosphate group as described herein. In some embodiments, the backbone modification of the phosphate backbone can include alterations that result in either an uncharged linker or a charged linker with unsymmetrical charge distribution. [00305] Examples of modified phosphate groups include, phosphorothioate, phosphoroselenates, borano phosphates, borano phosphate esters, hydrogen phosphonates, phosphoroamidates, alkyl or aryl phosphonates and phosphotriesters. The phosphorous atom in an unmodified phosphate group is achiral. However, replacement of one of the non-bridging oxygens with one of the above atoms or groups of atoms can render the phosphorous atom chiral. The stereogenic phosphorous atom can possess either the “R” configuration (herein Rp) or the “S” configuration (herein Sp). The backbone can also be modified by replacement of a bridging oxygen, (i.e., the oxygen that links the phosphate to the nucleoside), with nitrogen (bridged phosphoroamidates), sulfur (bridged phosphorothioates) and carbon (bridged methylenephosphonates). The replacement can occur at either linking oxygen or at both of the linking oxygens. [00306] The phosphate group can be replaced by non-phosphorus containing connectors in certain backbone modifications. In some embodiments, the charged phosphate group can be replaced by a neutral moiety. Examples of moieties which can replace the phosphate group can include, without limitation, e.g., methyl phosphonate, hydroxylamino, siloxane, carbonate,
Attorney Docket: 01155-0064-00PCT carboxymethyl, carbamate, amide, thioether, ethylene oxide linker, sulfonate, sulfonamide, thioformacetal, formacetal, oxime, methyleneimino, methylenemethylimino, methylenehydrazo, methylenedimethylhydrazo and methyleneoxymethylimino. [00307] In some embodiments, the disclosure comprises a sgRNA comprising one or more modifications within one or more of the following regions: the nucleotides at the 5' terminus; the lower stem region; the bulge region; the upper stem region; the nexus region; the hairpin 1 region; the hairpin 2 region; and the nucleotides at the 3' terminus. In some embodiments, the modification comprises a 2'-O-methyl (2'-O-Me) modified nucleotide. In some embodiments, the modification comprises a 2'-fluoro (2'-F) modified nucleotide. In some embodiments, the modification comprises a phosphorothioate (PS) bond between nucleotides. [00308] In some embodiments, the first three or four nucleotides at the 5' terminus, and the last three or four nucleotides at the 3' terminus are modified. In some embodiments, the first four nucleotides at the 5' terminus, and the last four nucleotides at the 3' terminus are linked with phosphorothioate (PS) bonds. In some embodiments, the modification comprises 2'-O- Me. In some embodiments, the modification comprises 2'-F. [00309] In some embodiments, the first four nucleotides at the 5' terminus and the last four nucleotides at the 3' terminus are linked with a PS bond, and the first three nucleotides at the 5' terminus and the last three nucleotides at the 3' terminus comprise 2'-O-Me modifications. [00310] In some embodiments, the first four nucleotides at the 5' terminus and the last four nucleotides at the 3' terminus are linked with a PS bond, and the first three nucleotides at the 5' terminus and the last three nucleotides at the 3' terminus comprise 2'-F modifications. [00311] In some embodiments, the sgRNA comprises the modification pattern of: (mN*mN*mN*NNNNNNNNNNNNNNNNNGUUUUAGAmGmCmUmAmGmAmAmAm UmAmGmCAAGUUAAAAUAAGGCUAGUCCGUUAUCAmAmCmUmUmGmAmAmA mAmAmGmUmGmGmCmAmCmCmGmAmGmUmCmGmGmUmGmCmU*mU*mU*mU (SEQ ID NO: 28), where N is any natural or non-natural nucleotide. A, C, G, and U are an adenine nucleotide, a cytidine nucleotide, a guanine nucleotide, and a uridine nucleotide, respectively. In certain embodiments, A, C, G, and U are each independently a naturally or non-naturally occurring nucleotide with the indicated base. In certain embodiments, A, C, G, and U are RNA nucleotides. In some embodiments, the sgRNA comprises the sequence disclosed in the sentence preceding this one. In some embodiments, the sgRNA comprises 2’O- methyl modification of the first three residues at its 5’ end, with phosphorothioate linkages between residues 1-2, 2-3, and 3-4 of the RNA.
Attorney Docket: 01155-0064-00PCT C. Template Nucleic Acid [00312] The compositions and methods disclosed herein may include a donor nucleic acid, i.e., a template nucleic acid. The template may be used to alter or insert a nucleic acid sequence at or near a target site for a Cas nuclease. In some embodiments, the methods comprise introducing a template to the cell. In some embodiments, a single template may be provided. In other embodiments, two or more templates may be provided such that editing may occur at two or more target sites. For example, different templates may be provided to edit a single gene in a cell, or two different genes in a cell. [00313] In some embodiments, the template may be used in homologous recombination. In some embodiments, the homologous recombination may result in the integration of the template sequence or a portion of the template sequence into the target nucleic acid molecule. In some embodiments, the template comprises regions having homology with corresponding regions of a T cell receptor sequence. In some embodiments, the template comprises regions having homology with corresponding regions of a TRAC locus, a B2M locus, an AAVS1 locus, and/or CIITA locus, or optionally a TRBC locus. In some embodiments, the template comprises regions having homology with corresponding regions of a TRAC locus. In some embodiments, the template comprises regions having homology with corresponding regions of a B2M locus. In some embodiments, the template comprises regions having homology with corresponding regions of a AAVS1 locus. In some embodiments, the template comprises regions having homology with corresponding regions of a CIITA locus. In some embodiments, the template comprises regions having homology with corresponding regions of a TRBC locus. In other embodiments, the template may be used in homology-directed repair, which involves DNA strand invasion at the site of the cleavage in the nucleic acid. In some embodiments, the homology-directed repair may result in including the template sequence in the edited target nucleic acid molecule. In other embodiments, the template may be used in gene editing mediated by blunt end insertion. In yet other embodiments, the template may be used in gene editing mediated by non-homologous end joining. In some embodiments, the template sequence has no similarity to the nucleic acid sequence near the cleavage site. In some embodiments, the template or a portion of the template sequence is incorporated. In some embodiments, the template includes flanking nucleic acid regions homologous to all or part of the target sequence. In some embodiments, the template includes flanking inverted terminal repeat (ITR) sequences.
Attorney Docket: 01155-0064-00PCT [00314] In some embodiments, the template may comprise a first homology arm and a second homology arm (also called a first and second nucleotide sequence) that are complementary to sequences located upstream and downstream of the cleavage site, respectively. Where a template contains two homology arms, each arm can be the same length or different lengths, and the sequence between the homology arms can be substantially similar or identical to the target sequence between the homology arms, or it can be entirely unrelated. In some embodiments, the degree of complementarity or percent identity between the first nucleotide sequence on the template and the sequence upstream of the cleavage site, and between the second nucleotide sequence on the template and the sequence downstream of the cleavage site, may permit homologous recombination, such as, e.g., high-fidelity homologous recombination, between the template and the target nucleic acid molecule. In some embodiments, the degree of complementarity may be about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98%, 99%, or 100%. In some embodiments, the degree of complementarity may be about 95%, 97%, 98%, 99%, or 100%. In some embodiments, the degree of complementarity may be at least 98%, 99%, or 100%. In some embodiments, the degree of complementarity may be 100%. In some embodiments, the percent identity may be about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98%, 99%, or 100%. In some embodiments, the percent identity may be about 95%, 97%, 98%, 99%, or 100%. In some embodiments, the percent identity may be at least 98%, 99%, or 100%. In some embodiments, the percent identity may be 100%. [00315] In some embodiments, the template sequence may correspond to, comprise, or consist of an endogenous sequence of a target cell. It may also or alternatively correspond to, comprise, or consist of an exogenous sequence of a target cell. As used herein, the term “endogenous sequence” refers to a sequence that is native to the cell. The term “exogenous sequence” refers to a sequence that is not native to a cell, or a sequence whose native location in the genome of the cell is in a different location. In some embodiments, the endogenous sequence may be a genomic sequence of the cell. In some embodiments, the endogenous sequence may be a chromosomal or extrachromosomal sequence. In some embodiments, the endogenous sequence may be a plasmid sequence of the cell. In some embodiments, the template sequence may be substantially identical to a portion of the endogenous sequence in a cell at or near the cleavage site, but comprise at least one nucleotide change. In some embodiments, editing the cleaved target nucleic acid molecule with the template may result in a mutation comprising an insertion, deletion, or substitution of one or more nucleotides of the
Attorney Docket: 01155-0064-00PCT target nucleic acid molecule. In some embodiments, the mutation may result in one or more amino acid changes in a protein expressed from a gene comprising the target sequence. [00316] In some embodiments, the mutation may result in one or more nucleotide changes in an RNA expressed from the target insertion site. In some embodiments, the mutation may alter the expression level of a target gene. In some embodiments, the mutation may result in increased or decreased expression of the target gene. In some embodiments, the mutation may result in gene knock-down. In some embodiments, the mutation may result in gene knock-out. In some embodiments, the mutation may result in restored gene function. In some embodiments, editing of the cleaved target nucleic acid molecule with the template may result in a change in an exon sequence, an intron sequence, a regulatory sequence, a transcriptional control sequence, a translational control sequence, a splicing site, or a non-coding sequence of the target nucleic acid molecule, such as DNA. [00317] In other embodiments, the template sequence may comprise an exogenous sequence. In some embodiments, the exogenous sequence may comprise a coding sequence. In some embodiments, the exogenous sequence may comprise a protein or RNA coding sequence (e.g., an ORF) operably linked to an exogenous promoter sequence such that, upon integration of the exogenous sequence into the target nucleic acid molecule, the cell is capable of expressing the protein or RNA encoded by the integrated sequence. In other embodiments, upon integration of the exogenous sequence into the target nucleic acid molecule, the expression of the integrated sequence may be regulated by an endogenous promoter sequence. In some embodiments, the exogenous sequence may provide a cDNA sequence encoding a protein or a portion of the protein. In yet other embodiments, the exogenous sequence may comprise or consist of an exon sequence, an intron sequence, a regulatory sequence, a transcriptional control sequence, a translational control sequence, a splicing site, or a non- coding sequence. In some embodiments, the integration of the exogenous sequence may result in restored gene function. In some embodiments, the integration of the exogenous sequence may result in a gene knock-in. In some embodiments, the integration of the exogenous sequence may result in a gene knock-out. [00318] The template may be of any suitable length. In some embodiments, the template may comprise 10, 15, 20, 25, 50, 75, 100, 150, 200, 500, 1000, 1500, 2000, 2500, 3000, 3500, 4000, 4500, 5000, 5500, 6000, or more nucleotides in length. The template may be a single- stranded nucleic acid. The template can be double-stranded or partially double-stranded nucleic acid. In some embodiments, the single stranded template is 20, 30, 40, 50, 75, 100, 125, 150, 175, or 200 nucleotides in length. In some embodiments, the template may comprise
Attorney Docket: 01155-0064-00PCT a nucleotide sequence that is complementary to a portion of the target nucleic acid molecule comprising the target sequence (i.e., a “homology arm”). In some embodiments, the template may comprise a homology arm that is complementary to the sequence located upstream or downstream of the cleavage site on the target nucleic acid molecule. In some embodiments, the template (i.e., donor nucleic acid) contains ssDNA or dsDNA containing flanking invert-terminal repeat (ITR) sequences. In some embodiments, the template is provided as a vector, plasmid, minicircle, nanocircle, or PCR product. In some embodiments, the donor nucleic acid comprises a vector. In some embodiments, the donor nucleic acid comprises a lentiviral vector. In some embodiments, the donor nucleic acid comprises a retroviral vector. In some embodiments, the donor nucleic acid comprises an adeno-associated virus (AAV) vector. D. Purification of Nucleic Acids [00319] In some embodiments, the nucleic acid is purified. In some embodiments, the nucleic acid is purified using a precipitation method (e.g., LiCl precipitation, alcohol precipitation, or an equivalent method, e.g., as described herein). In some embodiments, the nucleic acid is purified using a chromatography-based method, such as an HPLC-based method or an equivalent method (e.g., as described herein). In some embodiments, the nucleic is purified using both a precipitation method (e.g., LiCl precipitation) and an HPLC-based method. E. Target Sequences [00320] In some embodiments, a CRISPR/Cas system of the present disclosure may be directed to and cleave a target sequence on a target nucleic acid molecule. For example, the target sequence may be recognized and cleaved by the Cas nuclease. In some embodiments, a target sequence for a Cas nuclease is located near the nuclease’s cognate PAM sequence. In some embodiments, a Class 2 Cas nuclease may be directed by a gRNA to a target sequence of a target nucleic acid molecule, where the gRNA hybridizes with and the Class 2 Cas protein cleaves the target sequence. In some embodiments, the guide RNA hybridizes with and a Class 2 Cas nuclease cleaves the target sequence adjacent to or comprising its cognate PAM. In some embodiments, the target sequence may be complementary to the targeting sequence of the guide RNA. In some embodiments, the degree of complementarity between a targeting sequence of a guide RNA and the portion of the corresponding target sequence that hybridizes to the guide RNA may be about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%,
Attorney Docket: 01155-0064-00PCT 98%, 99%, or 100%. In some embodiments, the percent identity between a targeting sequence of a guide RNA and the portion of the corresponding target sequence that hybridizes to the guide RNA may be about 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98%, 99%, or 100%. In some embodiments, the homology region of the target is adjacent to a cognate PAM sequence. In some embodiments, the target sequence may comprise a sequence 100% complementary with the targeting sequence of the guide RNA. In other embodiments, the target sequence may comprise at least one mismatch, deletion, or insertion, as compared to the targeting sequence of the guide RNA. [00321] The length of the target sequence may depend on the nuclease system used. For example, the targeting sequence of a guide RNA for a CRISPR/Cas system may comprise 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, or more than 50 nucleotides in length and the target sequence is a corresponding length, optionally adjacent to a PAM sequence. In some embodiments, the target sequence may comprise 15-24 nucleotides in length. In some embodiments, the target sequence may comprise 17-21 nucleotides in length. In some embodiments, the target sequence may comprise 20 nucleotides in length. When nickases are used, the target sequence may comprise a pair of target sequences recognized by a pair of nickases that cleave opposite strands of the DNA molecule. In some embodiments, the target sequence may comprise a pair of target sequences recognized by a pair of nickases that cleave the same strands of the DNA molecule. In some embodiments, the target sequence may comprise a part of target sequences recognized by one or more Cas nucleases. [00322] The target nucleic acid molecule may be any DNA or RNA molecule that is endogenous or exogenous to a cell. In some embodiments, the target nucleic acid molecule may be an episomal DNA, a plasmid, a genomic DNA, viral genome, mitochondrial DNA, or chromosomal DNA from a cell or in the cell. In some embodiments, the target sequence of the target nucleic acid molecule may be a genomic sequence from a cell or in a cell, including a human cell. [00323] In further embodiments, the target sequence may be a viral sequence. In further embodiments, the target sequence may be a pathogen sequence. In yet other embodiments, the target sequence may be a synthesized sequence. In further embodiments, the target sequence may be a chromosomal sequence. In certain embodiments, the target sequence may comprise a translocation junction, e.g., a translocation associated with a cancer. In some embodiments, the target sequence may be on a eukaryotic chromosome, such as a human chromosome.
Attorney Docket: 01155-0064-00PCT [00324] In some embodiments, the target sequence may be located in a coding sequence of a gene, an intron sequence of a gene, a regulatory sequence, a transcriptional control sequence of a gene, a translational control sequence of a gene, a splicing site or a non-coding sequence between genes. In some embodiments, the gene may be a protein coding gene. In other embodiments, the gene may be a non-coding RNA gene. In some embodiments, the target sequence may comprise all or a portion of a disease-associated gene. In some embodiments, the target sequence may be located in a non-genic functional site in the genome, for example a site that controls aspects of chromatin organization, such as a scaffold site or locus control region. [00325] In some embodiments involving a Cas nuclease, such as a Class 2 Cas nuclease, the target sequence may be adjacent to a protospacer adjacent motif (“PAM”). In some embodiments, the PAM may be adjacent to or within 1, 2, 3, or 4, nucleotides of the 3' end of the target sequence. The length and the sequence of the PAM may depend on the Cas protein used. For example, the PAM may be selected from a consensus or a particular PAM sequence for a specific Cas9 protein or Cas9 ortholog, including those disclosed in Figure 1 of Ran et al., Nature, 520: 186-191 (2015), and Figure S5 of Zetsche 2015, the relevant disclosure of each of which is incorporated herein by reference. In some embodiments, the PAM may be 2, 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides in length. Non-limiting exemplary PAM sequences include NGG, NGGNG, NG, NAAAAN, NNAAAAW, NNNNACA, GNNNCNNA, TTN, and NNNNGATT (wherein N is defined as any nucleotide, and W is defined as either A or T). In some embodiments, the PAM sequence may be NGG. In some embodiments, the PAM sequence may be NGGNG. In some embodiments, the PAM sequence may be TTN. In some embodiments, the PAM sequence may be NNAAAAW. VI. Exemplary Lipid Nucleic Acid Assemblies [00326] Disclosed herein are various embodiments using lipid nucleic acid assemblies comprising genome editing tools, such as RNAs, including CRISPR/Cas components and RNAs that express the same. [00327] As used herein, “lipid nucleic acid assembly composition” refers to lipid-based delivery compositions, including lipid nanoparticles (LNPs) and lipoplexes. In some embodiments, “LNP compositions” are used interchangeably with “LNPs” or “LNP.” [00328] In some embodiments, LNP refers to lipid nanoparticles with a diameter of <100 nm, or a population of LNP with an average diameter of <100 nm, as measured by dynamic light scattering. In some embodiments, the particle size is a number average. In some
Attorney Docket: 01155-0064-00PCT embodiments, the particle size is a Z-average. In certain embodiments, an LNP has a diameter of about 1-250 nm, 10-200 nm, about 20-150 nm, about 35-150 nm, about 50-150 nm, about 50-100 nm, about 50-120 nm, about 60-100 nm, about 75-150 nm, about 75-120 nm, or about 75-100 nm, or a population of the LNP with an average diameter, as measured by dynamic light scattering, of about 10-200 nm, about 20-150 nm, about 35-150 nm, about 50-150 nm, about 50-100 nm, about 50-120 nm, about 60-100 nm, about 75-150 nm, about 75-120 nm, or about 75-100 nm. In preferred embodiments, an LNP composition has a diameter of 75-150 nm. [00329] LNPs are formed by precise mixing a lipid component (e.g., in ethanol) with an aqueous nucleic acid component and LNPs are uniform in size. Lipoplexes are particles formed by bulk mixing the lipid and nucleic acid components and are between about 100nm and 1 micron in size. In certain embodiments the lipid nucleic acid assemblies are LNPs. As used herein, a “lipid nucleic acid assembly” comprises a plurality of (i.e. more than one) lipid molecules physically associated with each other by intermolecular forces. A lipid nucleic acid assembly may comprise a bioavailable lipid having a pKa value of <7.5 or <7. The lipid nucleic acid assemblies are formed by mixing an aqueous nucleic acid-containing solution with an organic solvent-based lipid solution, e.g., 100% ethanol. Suitable solutions or solvents include or may contain: water, PBS, Tris buffer, NaCl, citrate buffer, ethanol, chloroform, diethylether, cyclohexane, tetrahydrofuran, methanol, isopropanol. A pharmaceutically acceptable buffer may optionally be comprised in a pharmaceutical formulation comprising the lipid nucleic acid assemblies, e.g., for an ex vivo ACT therapy. In some embodiments, the aqueous solution comprises an RNA, such as an mRNA or a gRNA. In some embodiments, the aqueous solution comprises an mRNA encoding an RNA-guided DNA binding agent, such as Cas9. [00330] In some embodiments, the lipid nucleic acid assembly formulations include an “amine lipid” (sometimes herein or elsewhere described as an “ionizable lipid” or a “biodegradable lipid”), together with an optional “helper lipid”, a “neutral lipid”, and a stealth lipid such as a PEG lipid. In some embodiments, the amine lipids or ionizable lipids are cationic depending on the pH. A. Amine Lipids [00331] In some embodiments, lipid nucleic acid assembly compositions comprise an “amine lipid”, which is, for example an ionizable lipid such as Lipid A, or Lipid D or their equivalents, including acetal analogs of Lipid A or Lipid D.
Attorney Docket: 01155-0064-00PCT [00332] In some embodiments, the amine lipid is Lipid A, which is (9Z,12Z)-3-((4,4- bis(octyloxy)butanoyl)oxy)-2-((((3-(diethylamino)propoxy)carbonyl)oxy)methyl)propyl octadeca-9,12-dienoate, also called 3-((4,4-bis(octyloxy)butanoyl)oxy)-2-((((3- (diethylamino)propoxy)carbonyl)oxy)methyl)propyl (9Z,12Z)-octadeca-9,12-dienoate. Lipid A can be depicted as: . pp. 84-86). In

some embodiments, the amine lipid is Lipid A, or an amine lipid provided in WO2020/219876, which is hereby incorporated by reference. [00334] In some embodiments, an amine lipid is an analog of Lipid A. In some embodiments, a Lipid A analog is an acetal analog of Lipid A. In particular lipid nucleic acid assembly compositions, the acetal analog is a C4-C12 acetal analog. In some embodiments, the acetal analog is a C5-C12 acetal analog. In additional embodiments, the acetal analog is a C5- C10 acetal analog. In further embodiments, the acetal analog is chosen from a C4, C5, C6, C7, C9, C10, C11, and C12 acetal analog. [00335] In some embodiments, the amine lipid is a compound having a structure of Formula IA , wherein

X1A is O, NH, or a direct bond; X2A is C2-3 alkylene; R3A is C1-3 alkyl; R2A is C1-3 alkyl, or R2A taken together with the nitrogen atom to which it is attached and 2-3 carbon atoms of X2A form a 5- or 6-membered ring, or
Attorney Docket: 01155-0064-00PCT R2A taken together with R3A and the nitrogen atom to which they are attached form a 5- membered ring; Y1A is C6-10 alkylene; Y2A is selected ; R4A is C4-11 alkyl; Z1A is C2-5
and
or or a salt thereof. [00336] In some embodiments, the amine lipid is a compound of Formula (IIA) , wherein
X1A is O, NH, or a direct bond; X2A is C2-3 alkylene; Z1A is C3 alkylene and R5A and R6A are each C6 alkyl, or Z1A is a direct bond and R5A and R6A are each C8 alkoxy; and ;
[00337] In certain embodiments, X1A is O. In other embodiments, X1A is NH. In still other embodiments, X1A is a direct bond. [00338] In certain embodiments, X2A is C3 alkylene. In particular embodiments, X2A is C2 alkylene. [00339] In certain embodiments, Z1A is a direct bond and R5A and R6A are each C8 alkoxy. In other embodiments, Z1A is C3 alkylene and R5A and R6A are each C6 alkyl.
Attorney Docket: 01155-0064-00PCT [00340] In certain embodiments, R8A is . In other
. [00341] lipid is a salt. [00342]
(IA) include: Compound Compound Number
Attorney Docket: 01155-0064-00PCT
Attorney Docket: 01155-0064-00PCT
Attorney Docket: 01155-0064-00PCT
[0033] n some embodments, te amne pd s pd , w c s nony 8-((7,7- bis(octyloxy)heptyl)(2-hydroxyethyl)amino)octanoate:
2018, 26(6), 1509-1519 (“Sabnis”), which are incorporated by reference in their entireties. In some embodiments,
Lipid D, or an amine lipid provided in WO2020072605, which is hereby incorporated by reference. [00345] In some embodiments, the amine lipid is a compound having a structure of Formula IB:
Attorney Docket: 01155-0064-00PCT wherein X
1B is C al
6-7 kylene; is not alkoxy;
- - - R
1B is C7-9 unbranched alkyl; and each R
2B is independently C
8 alkyl or C
8 alkoxy; or a salt thereof [00346] In some embodiments, the amine lipid is a compound of Formula (IIB) wherein
X
1B is C
6-7 alkylene; Z
1B is C2-3 alkylene; R
1B is C
7-9 unbranched alkyl; and each R
2B is C8 alkyl; or a salt thereof. [00347] In certain embodiments, X
1B is C6 alkylene. In other embodiments, X
1B is C7 alkylene. [00348] In certain embodiments, Z
1B is a direct bond and R
5B and R
6B are each C8 alkoxy. In other embodiments, Z
1B is C
3 alkylene and R
5B and R
6B are each C
6 alkyl. [00349] In certain embodiments, X
2B is and R
2B is not alkoxy. In other embodiments, X
2B is absent.
Attorney Docket: 01155-0064-00PCT [00350] In certain embodiments, Z
1B is C2 alkylene; In other embodiments, Z
1B is C3 alkylene. [00351] In certain embodiments, Z
2B is -OH. In other embodiments, Z
2B is - NHC(=O)OCH
3. In other embodiments, Z
2B is -NHS(=O)
2CH
3. [00352] In certain embodiments, R
1B is C7 unbranched alkylene. In other embodiments, R
1B is C
8 branched or unbranched alkylene. In other embodiments, R
1B is C
9 branched or unbranched alkylene. [00353] In certain embodiments, the amine lipid is a salt. [00354] Representative compounds of Formula (IB) include: Compound Compound Number
Attorney Docket: 01155-0064-00PCT
eic acid assemblies described herein are biodegradable in vivo or ex vivo. The amine lipids have low toxicity (e.g., are tolerated in animal models without adverse effect in amounts of greater than or equal to 10 mg/kg). In some embodiments, lipid nucleic acid assemblies comprising an amine lipid include those where at least 75% of the amine lipid is cleared from the plasma or the engineered cell within 8, 10, 12, 24, or 48 hours, or 3, 4, 5, 6, 7, or 10 days. In some embodiments, lipid nucleic acid assemblies comprising an amine lipid include those where at least 50% of the nucleic acid, e.g., mRNA or gRNA, is cleared from the plasma within 8, 10, 12, 24, or 48 hours, or 3, 4, 5, 6, 7, or 10 days. In some embodiments, lipid nucleic acid assemblies comprising an amine lipid include those where at least 50% of the lipid nucleic acid assembly is cleared from the plasma within 8, 10, 12, 24, or 48 hours, or 3, 4, 5, 6, 7, or 10 days, for example by measuring a lipid (e.g. an amine lipid), nucleic acid, e.g., RNA/mRNA, or other component. In some embodiments, lipid-encapsulated versus free lipid, RNA, or nucleic acid component of the lipid nucleic acid assembly is measured. [00356] Biodegradable lipids include, for example the biodegradable lipids of WO/2020/219876 (e.g., at pp.13-33, 66-87), WO/2020/118041, WO/2020/072605 (e.g., at pp. 5-12, 21-29, 61-68, WO/2019/067992, WO/2017/173054, WO2015/095340, and WO2014/136086, and LNPs include LNP compositions described therein, the lipids and compositions of which are hereby incorporated by reference. [00357] Lipid clearance may be measured as described in literature. See Maier, M.A., et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther. 2013, 21(8), 1570-78 (“Maier”). For example, in Maier,
Attorney Docket: 01155-0064-00PCT LNP-siRNA systems containing luciferases-targeting siRNA were administered to six- to eight-week old male C57Bl/6 mice at 0.3 mg/kg by intravenous bolus injection via the lateral tail vein. Blood, liver, and spleen samples were collected at 0.083, 0.25, 0.5, 1, 2, 4, 8, 24, 48, 96, and 168 hours post-dose. Mice were perfused with saline before tissue collection and blood samples were processed to obtain plasma. All samples were processed and analyzed by LC- MS. Further, Maier describes a procedure for assessing toxicity after administration of LNP- siRNA formulations. For example, a luciferase-targeting siRNA was administered at 0, 1, 3, 5, and 10 mg/kg (5 animals/group) via single intravenous bolus injection at a dose volume of 5 mL/kg to male Sprague-Dawley rats. After 24 hours, about 1 mL of blood was obtained from the jugular vein of conscious animals and the serum was isolated. At 72 hours post-dose, all animals were euthanized for necropsy. Assessments of clinical signs, body weight, serum chemistry, organ weights and histopathology were performed. Although Maier describes methods for assessing siRNA-LNP formulations, these methods may be applied to assess clearance, pharmacokinetics, and toxicity of administration of lipid nucleic acid assembly compositions of the present disclosure. [00358] Ionizable and bioavailable lipids for LNP delivery of nucleic acids known in the art are suitable. Lipids may be ionizable depending upon the pH of the medium they are in. For example, in a slightly acidic medium, the lipid, such as an amine lipid, may be protonated and thus bear a positive charge. Conversely, in a slightly basic medium, such as, for example, blood where pH is approximately 7.35, the lipid, such as an amine lipid, may not be protonated and thus bear no charge. [00359] The ability of a lipid to bear a charge is related to its intrinsic pKa. In some embodiments, the amine lipids of the present disclosure may each, independently, have a pKa in the range of from about 5.1 to about 7.4. In some embodiments, the bioavailable lipids of the present disclosure may each, independently, have a pKa in the range of from about 5.1 to about 7.4, such as from about 5.5 to about 6.6, from about 5.6 to about 6.4, from about 5.8 to about 6.2, or from about 5.8 to about 6.5. For example, the amine lipids of the present disclosure may each, independently, have a pKa in the range of from about 5.8 to about 6.5. Lipids with a pKa ranging from about 5.1 to about 7.4 are effective for delivery of cargo in vivo, e.g. to the liver. Further, it has been found that lipids with a pKa ranging from about 5.3 to about 6.4 are effective for delivery in vivo, e.g. to tumors. See, e.g., WO2014/136086.
Attorney Docket: 01155-0064-00PCT B. Additional Lipids [00360] “Neutral lipids” suitable for use in a lipid composition of the disclosure include, for example, a variety of neutral, uncharged or zwitterionic lipids. Examples of neutral phospholipids suitable for use in the present disclosure include, but are not limited to, 5- heptadecylbenzene-1,3-diol (resorcinol), dipalmitoylphosphatidylcholine (DPPC), distearoylphosphatidylcholine (DSPC), pohsphocholine (DOPC), dimyristoylphosphatidylcholine (DMPC), phosphatidylcholine (PLPC), 1,2-distearoyl-sn- glycero-3-phosphocholine (DAPC), phosphatidylethanolamine (PE), egg phosphatidylcholine (EPC), dilauryloylphosphatidylcholine (DLPC), dimyristoylphosphatidylcholine (DMPC), 1- myristoyl-2-palmitoyl phosphatidylcholine (MPPC), 1-palmitoyl-2-myristoyl phosphatidylcholine (PMPC), 1-palmitoyl-2-stearoyl phosphatidylcholine (PSPC), 1,2- diarachidoyl-sn-glycero-3-phosphocholine (DBPC), 1-stearoyl-2-palmitoyl phosphatidylcholine (SPPC), 1,2-dieicosenoyl-sn-glycero-3-phosphocholine (DEPC), palmitoyloleoyl phosphatidylcholine (POPC), lysophosphatidyl choline, dioleoyl phosphatidylethanolamine (DOPE), dilinoleoylphosphatidylcholine distearoylphosphatidylethanolamine (DSPE), dimyristoyl phosphatidylethanolamine (DMPE), dipalmitoyl phosphatidylethanolamine (DPPE), palmitoyloleoyl phosphatidylethanolamine (POPE), lysophosphatidylethanolamine and combinations thereof. In one embodiment, the neutral phospholipid may be selected from the group consisting of distearoylphosphatidylcholine (DSPC) and dimyristoyl phosphatidyl ethanolamine (DMPE). In another embodiment, the neutral phospholipid may be distearoylphosphatidylcholine (DSPC). [00361] “Helper lipids” include steroids, sterols, and alkyl resorcinols. Helper lipids suitable for use in the present disclosure include, but are not limited to, cholesterol, 5- heptadecylresorcinol, and cholesterol hemisuccinate. In one embodiment, the helper lipid may be cholesterol. In one embodiment, the helper lipid may be cholesterol hemisuccinate. [00362] “Stealth lipids” are lipids that alter the length of time the nanoparticles can exist in vivo (e.g., in the blood). Stealth lipids may assist in the formulation process by, for example, reducing particle aggregation and controlling particle size. Stealth lipids used herein may modulate pharmacokinetic properties of the lipid nucleic acid assembly or aid in stability of the nanoparticle ex vivo. Stealth lipids suitable for use in a lipid composition of the disclosure include, but are not limited to, stealth lipids having a hydrophilic head group linked to a lipid moiety. Stealth lipids suitable for use in a lipid composition of the present disclosure and
Attorney Docket: 01155-0064-00PCT information about the biochemistry of such lipids can be found in Romberg et al., Pharmaceutical Research, Vol. 25, No. 1, 2008, pg. 55-71 and Hoekstra et al., Biochimica et Biophysica Acta 1660 (2004) 41-52. Additional suitable PEG lipids are disclosed, e.g., in WO 2006/007712. [00363] In one embodiment, the hydrophilic head group of stealth lipid comprises a polymer moiety selected from polymers based on PEG. Stealth lipids may comprise a lipid moiety. In some embodiments, the stealth lipid is a PEG lipid. [00364] In one embodiment, a stealth lipid comprises a polymer moiety selected from polymers based on PEG (sometimes referred to as poly(ethylene oxide)), poly(oxazoline), poly(vinyl alcohol), poly(glycerol), poly(N-vinylpyrrolidone), polyaminoacids and poly[N-(2- hydroxypropyl)methacrylamide]. [00365] In one embodiment, the PEG lipid comprises a polymer moiety based on PEG (sometimes referred to as poly(ethylene oxide)). [00366] The PEG lipid further comprises a lipid moiety. In some embodiments, the lipid moiety may be derived from diacylglycerol or diacylglycamide, including those comprising a dialkylglycerol or dialkylglycamide group having alkyl chain length independently comprising from about C4 to about C40 saturated or unsaturated carbon atoms, wherein the chain may comprise one or more functional groups such as, for example, an amide or ester. In some embodiments, the alkyl chain length comprises about C10 to C20. The dialkylglycerol or dialkylglycamide group can further comprise one or more substituted alkyl groups. The chain lengths may be symmetrical or asymmetrical. [00367] Unless otherwise indicated, the term “PEG” as used herein means any polyethylene glycol or other polyalkylene ether polymer. In one embodiment, PEG is an optionally substituted linear or branched polymer of ethylene glycol or ethylene oxide. In one embodiment, PEG is unsubstituted. In one embodiment, the PEG is substituted, e.g., by one or more alkyl, alkoxy, acyl, hydroxy, or aryl groups. In one embodiment, the term includes PEG copolymers such as PEG-polyurethane or PEG-polypropylene (see, e.g., J. Milton Harris, Poly(ethylene glycol) chemistry: biotechnical and biomedical applications (1992)); in another embodiment, the term does not include PEG copolymers. In one embodiment, the PEG has a molecular weight of from about 130 to about 50,000, in a sub-embodiment, about 150 to about 30,000, in a sub-embodiment, about 150 to about 20,000, in a sub-embodiment about 150 to about 15,000, in a sub-embodiment, about 150 to about 10,000, in a sub-embodiment, about 150 to about 6,000, in a sub-embodiment, about 150 to about 5,000, in a sub-embodiment, about 150 to about 4,000, in a sub-embodiment, about 150 to about 3,000, in a sub-
Attorney Docket: 01155-0064-00PCT embodiment, about 300 to about 3,000, in a sub-embodiment, about 1,000 to about 3,000, and in a sub-embodiment, about 1,500 to about 2,500. [00368] In some embodiments, the PEG (e.g., conjugated to a lipid moiety or lipid, such as a stealth lipid), is a “PEG-2K,” also termed “PEG 2000,” which has an average molecular weight of about 2,000 Daltons. PEG-2K is represented herein by the following formula (IV), wherein n is 45, meaning that the number averaged degree of polymerization comprises about 45 subunits . However, other PEG embodiments known in the art may be used,

the number-averaged degree of polymerization comprises about 23 subunits (n=23), and/or 68 subunits (n=68). In some embodiments, n may range from about 30 to about 60. In some embodiments, n may range from about 35 to about 55. In some embodiments, n may range from about 40 to about 50. In some embodiments, n may range from about 42 to about 48. In some embodiments, n may be 45. In some embodiments, R may be selected from H, substituted alkyl, and unsubstituted alkyl. In some embodiments, R may be unsubstituted alkyl. In some embodiments, R may be methyl. [00369] In any of the embodiments described herein, the PEG lipid may be selected from PEG-dilauroylglycerol, PEG-dimyristoylglycerol (PEG-DMG) (catalog # GM-020 from NOF, Tokyo, Japan), PEG-dipalmitoylglycerol, PEG-distearoylglycerol (PEG-DSPE) (catalog # DSPE-020CN, NOF, Tokyo, Japan), PEG-dilaurylglycamide, PEG-dimyristylglycamide, PEG-dipalmitoylglycamide, and PEG-distearoylglycamide, PEG-cholesterol (1-[8'-(Cholest- 5-en-3[beta]-oxy)carboxamido-3',6'-dioxaoctanyl]carbamoyl-[omega]-methyl-poly(ethylene glycol), PEG-DMB (3,4-ditetradecoxylbenzyl-[omega]-methyl-poly(ethylene glycol)ether), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (PEG2k-DMG) (cat. #880150P from Avanti Polar Lipids, Alabaster, Alabama, USA), 1,2- distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (PEG2k-DSPE) (cat. #880120C from Avanti Polar Lipids, Alabaster, Alabama, USA), 1,2- distearoyl-sn-glycerol, methoxypolyethylene glycol (PEG2k-DSG; GS-020, NOF Tokyo, Japan), poly(ethylene glycol)-2000-dimethacrylate (PEG2k-DMA), and 1,2- distearyloxypropyl-3-amine-N-[methoxy(polyethylene glycol)-2000] (PEG2k-DSA). In one embodiment, the PEG lipid may be PEG2k-DMG. In some embodiments, the PEG lipid may be PEG2k-DSG. In one embodiment, the PEG lipid may be PEG2k-DSPE. In one embodiment, the PEG lipid may be PEG2k-DMA. In one embodiment, the PEG lipid may be PEG2k-C-DMA. In one embodiment, the PEG lipid may be compound S027, disclosed in
Attorney Docket: 01155-0064-00PCT WO2016/010840 (paragraphs [00240] to [00244]). In one embodiment, the PEG lipid may be PEG2k-DSA. In one embodiment, the PEG lipid may be PEG2k-C11. In some embodiments, the PEG lipid may be PEG2k-C14. In some embodiments, the PEG lipid may be PEG2k-C16. In some embodiments, the PEG lipid may be PEG2k-C18. C. Lipid Nucleic Acid Assembly Compositions [00370] The lipid nucleic acid assembly may contain (i) a biodegradable lipid, (ii) an optional neutral lipid, (iii) a helper lipid, and (iv) a stealth lipid, such as a PEG lipid. The lipid nucleic acid assembly may contain a biodegradable lipid and one or more of a neutral lipid, a helper lipid, and a stealth lipid, such as a PEG lipid. [00371] The lipid nucleic acid assembly may contain (i) an amine lipid for encapsulation and for endosomal escape, (ii) a neutral lipid for stabilization, (iii) a helper lipid, also for stabilization, and (iv) a stealth lipid, such as a PEG lipid. The lipid nucleic acid assembly may contain an amine lipid and one or more of a neutral lipid, a helper lipid, also for stabilization, and a stealth lipid, such as a PEG lipid. [00372] A lipid nucleic acid assembly composition may comprise a nucleic acid, e.g., an RNA, component that includes one or more of an RNA-guided DNA-binding agent, a Cas nuclease mRNA, a Class 2 Cas nuclease mRNA, a Cas9 mRNA, and a gRNA. In some embodiments, a lipid nucleic acid assembly composition may include a Class 2 Cas nuclease and a gRNA as the RNA component. In some embodiments, n lipid nucleic acid assembly composition may comprise the RNA component, an amine lipid, a helper lipid, a neutral lipid, and a stealth lipid. In certain lipid nucleic acid assembly compositions, the helper lipid is cholesterol. In other compositions, the neutral lipid is DSPC. In additional embodiments, the stealth lipid is PEG2k-DMG or PEG2k-C11. In some embodiments, the lipid nucleic acid assembly composition comprises Lipid A or an equivalent of Lipid A; a helper lipid; a neutral lipid; a stealth lipid; and an RNA such as a gRNA. In some embodiments, the lipid nucleic acid assembly composition comprises Lipid A or an equivalent of Lipid A; a helper lipid; a stealth lipid; and an RNA such as a gRNA. In some compositions, the amine lipid is Lipid A. In some compositions, the amine lipid is Lipid A or an acetal analog thereof; the helper lipid is cholesterol; the neutral lipid is DSPC; and the stealth lipid is PEG2k-DMG. [00373] In some embodiments, lipid compositions are described according to the respective molar ratios of the component lipids in the formulation. Embodiments of the present disclosure provide lipid compositions described according to the respective molar ratios of the component lipids in the formulation. In one embodiment, the mol % of the amine lipid may be from about
Attorney Docket: 01155-0064-00PCT 30 mol % to about 60 mol %. In one embodiment, the mol % of the amine lipid may be from about 40 mol % to about 60 mol %. In one embodiment, the mol % of the amine lipid may be from about 45 mol % to about 60 mol %. In one embodiment, the mol % of the amine lipid may be from about 50 mol % to about 60 mol %. In one embodiment, the mol % of the amine lipid may be from about 55 mol % to about 60 mol %. In one embodiment, the mol % of the amine lipid may be from about 50 mol % to about 55 mol %. In one embodiment, the mol % of the amine lipid may be about 50 mol %. In one embodiment, the mol % of the amine lipid may be about 55 mol %. In some embodiments, the amine lipid mol % of the lipid nucleic acid assembly batch will be ±30%, ±25%, ±20%, ±15%, ±10%, ±5%, or ±2.5% of the target mol %. In some embodiments, the amine lipid mol % of the lipid nucleic acid assembly batch will be ±4 mol %, ±3 mol %, ±2 mol %, ±1.5 mol %, ±1 mol %, ±0.5 mol %, or ±0.25 mol % of the target mol %. All mol % numbers are given as a fraction of the lipid component of the lipid nucleic acid assembly compositions. In some embodiments, lipid nucleic acid assembly inter- lot variability of the amine lipid mol % will be less than 15%, less than 10% or less than 5%. [00374] In one embodiment, the mol % of the neutral lipid may be from about 5 mol % to about 15 mol %. In one embodiment, the mol % of the neutral lipid may be from about 7 mol % to about 12 mol %. In one embodiment, the mol % of the neutral lipid may be about 9 mol %. In some embodiments, the neutral lipid mol % of the lipid nucleic acid assembly batch will be ±30%, ±25%, ±20%, ±15%, ±10%, ±5%, or ±2.5% of the target neutral lipid mol %. In some embodiments, lipid nucleic acid assembly inter-lot variability will be less than 15%, less than 10% or less than 5%. [00375] In one embodiment, the mol % of the helper lipid may be from about 20 mol % to about 60 mol %. In one embodiment, the mol % of the helper lipid may be from about 25 mol % to about 55 mol %. In one embodiment, the mol % of the helper lipid may be from about 25 mol % to about 50 mol %. In one embodiment, the mol % of the helper lipid may be from about 25 mol % to about 40 mol %. In one embodiment, the mol % of the helper lipid may be from about 30 mol % to about 50 mol %. In one embodiment, the mol % of the helper lipid may be from about 30 mol % to about 40 mol %. In one embodiment, the mol % of the helper lipid is adjusted based on amine lipid, neutral lipid, and PEG lipid concentrations to bring the lipid component to 100 mol %. In some embodiments, the helper mol % of the lipid nucleic acid assembly batch will be ±30%, ±25%, ±20%, ±15%, ±10%, ±5%, or ±2.5% of the target mol %. In some embodiments, lipid nucleic acid assembly inter-lot variability will be less than 15%, less than 10% or less than 5%.
Attorney Docket: 01155-0064-00PCT [00376] In one embodiment, the mol % of the PEG lipid may be from about 1 mol % to about 10 mol %. In one embodiment, the mol % of the PEG lipid may be from about 2 mol % to about 10 mol %. In one embodiment, the mol % of the PEG lipid may be from about 1 mol % to about 3 mol %. In one embodiment, the mol % of the PEG lipid may be from about 2 mol % to about 4 mol %. In one embodiment, the mol % of the PEG lipid may be from about 1.5 mol % to about 2 mol %. In one embodiment, the mol % of the PEG lipid may be from about 2.5 mol % to about 4 mol %. In one embodiment, the mol % of the PEG lipid may be about 3 mol %. In one embodiment, the mol % of the PEG lipid may be about 2.5 mol %. In one embodiment, the mol % of the PEG lipid may be about 2 mol %. In one embodiment, the mol % of the PEG lipid may be about 1.5 mol %. In some embodiments, the PEG lipid mol % of the lipid nucleic acid assembly batch will be ±30%, ±25%, ±20%, ±15%, ±10%, ±5%, or ±2.5% of the target PEG lipid mol %. In some embodiments, lipid nucleic acid assembly composition, e.g. the LNP composition, inter-lot variability will be less than 15%, less than 10% or less than 5%. Embodiments of the present disclosure provide LNP compositions, for example, LNP compositions comprising an ionizable lipid (e.g., Lipid A or one of its analogs), a helper lipid, a helper lipid, and a PEG lipid, described according to the respective molar ratios of the component lipids in the formulation. In certain embodiments, the amount of the ionizable lipid is from about 25 mol % to about 45 mol %; the amount of the neutral lipid is from about 10 mol % to about 30 mol %; the amount of the helper lipid is from about 25 mol % to about 65 mol %; and the amount of the PEG lipid is from about 1.5 mol % to about 3.5 mol %. In certain embodiments, the amount of the ionizable lipid is from about 29-44 mol % of the lipid component; the amount of the neutral lipid is from about 11-28 mol % of the lipid component; the amount of the helper lipid is from about 28-55 mol % of the lipid component; and the amount of the PEG lipid is from about 2.3-3.5 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is from about 29-38 mol % of the lipid component; the amount of the neutral lipid is from about 11-20 mol % of the lipid component; the amount of the helper lipid is from about 43-55 mol % of the lipid component; and the amount of the PEG lipid is from about 2.3-2.7 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is from about 25-34 mol % of the lipid component; the amount of the neutral lipid is from about 10-20 mol % of the lipid component; the amount of the helper lipid is from about 45-65 mol % of the lipid component; and the amount of the PEG lipid is from about 2.5-3.5 mol % of the lipid component. In certain embodiments, the ionizable lipid is about 30-43 mol % of the lipid component; the amount of
Attorney Docket: 01155-0064-00PCT the neutral lipid is about 10-17 mol % of the lipid component; the amount of the helper lipid is about 43.5-56 mol % of the lipid component; and the amount of the PEG lipid is about 1.5-3 mol % of the lipid component. In certain embodiments, the ionizable lipid is about 33 mol % of the lipid component; the amount of the neutral lipid is about 15 mol % of the lipid component; the amount of the helper lipid is about 49 mol % of the lipid component; and the amount of the PEG lipid is about 3 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is about 32.9 mol % of the lipid component; the amount of the neutral lipid is about 15.2 mol % of the lipid component; the amount of the helper lipid is about 49.2 mol % of the lipid component; and the amount of the PEG lipid is about 2.7 mol % of the lipid component. [00377] In certain embodiments, the amount of the ionizable lipid (e.g., Lipid A or one of its analogs) is about 20-50 mol %, about 25-34 mol %, about 25-38 mol %, about 25-45 mol %, about 29-38 mol %, about 29-43 mol %, about 29-34 mol %, about 30-34 mol %, about 30- 38 mol %, about 30-43 mol %, about 30-43 mol %, or about 33 mol %. In certain embodiments, the amount of the neutral lipid is about 10-30 mol %, about 11-30 mol %, about 11-20 mol %, about 13-17 mol %, or about 15 mol %. In certain embodiments, the amount of the helper lipid is about 35-50 mol %, about 35-65 mol %, about 35-55 mol %, about 38-50 mol %, about 38- 55 mol %, about 38-65 mol %, about 40-50 mol %, about 40-65 mol %, about 43-65 mol %, about 43-55 mol %, or about 49 mol %. In certain embodiments, the amount of the PEG lipid is about 1.5-3.5 mol %, about 2.0-2.7 mol %, about 2.0-3.5 mol %, about 2.3-3.5 mol %, about 2.3-2.7 mol %, about 2.5-3.5 mol %, about 2.5-2.7 mol %, about 2.9-3.5 mol %, or about 2.7 mol %. [00378] Other embodiments of the present disclosure provide LNP compositions, for example, LNP compositions comprising an ionizable lipid (e.g., Lipid D or one of its analogs), a helper lipid, a helper lipid, and a PEG lipid, described according to the respective molar ratios of the component lipids in the formulation. In certain embodiments, the amount of the ionizable lipid is from about 25 mol % to about 50 mol %; the amount of the neutral lipid is from about 7 mol % to about 25 mol %; the amount of the helper lipid is from about 39 mol % to about 65 mol %; and the amount of the PEG lipid is from about 0.5 mol % to about 1.8 mol %. In certain embodiments, the amount of the ionizable lipid is from about 27-40 mol % of the lipid component; the amount of the neutral lipid is from about 10-20 mol % of the lipid component; the amount of the helper lipid is from about 50-60 mol % of the lipid component; and the amount of the PEG lipid is from about 0.9-1.6 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is from about 30-45 mol % of the lipid
Attorney Docket: 01155-0064-00PCT component; the amount of the neutral lipid is from about 10-15 mol % of the lipid component; the amount of the helper lipid is from about 39-59 mol % of the lipid component; and the amount of the PEG lipid is from about 1-1.5 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is from about 30-45 mol % of the lipid component; the amount of the neutral lipid is from about 10-15 mol % of the lipid component; the amount of the helper lipid is from about 39-59 mol % of the lipid component; and the amount of the PEG lipid is from about 1-1.5 mol % of the lipid component. In certain embodiments, the ionizable lipid is about 30 mol % of the lipid component; the amount of the neutral lipid is about 10 mol % of the lipid component; the amount of the helper lipid is about 59 mol % of the lipid component; and the amount of the PEG lipid is about 1-1.5 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is about 40 mol % of the lipid component; the amount of the neutral lipid is about 15 mol % of the lipid component; the amount of the helper lipid is about 43.5 mol % of the lipid component; and the amount of the PEG lipid is about 1.5 mol % of the lipid component. In certain embodiments, the amount of the ionizable lipid is about 50 mol % of the lipid component; the amount of the neutral lipid is about 10 mol % of the lipid component; the amount of the helper lipid is about 39 mol % of the lipid component; and the amount of the PEG lipid is about 1 mol % of the lipid component. [00379] In certain embodiments, the amount of the ionizable lipid (e.g., Lipid D or one of its analogs) is about 20-55 mol %, about 20-45 mol %, about 20-40 mol %, about 27-40 mol %, about 27-45 mol %, about 27-55 mol %, about 30-40 mol %, about 30-45 mol %, about 30- 55 mol %, about 30 mol %, about 40 mol %, or about 50 mol %. In certain embodiments, the amount of the neutral lipid is about 7-25 mol %, about 10-25 mol %, about 10-20 mol %, about 15-20 mol %, about 8-15 mol %, about 10-15 mol %, about 10 mol %, or about 15 mol %. In certain embodiments, the amount of the helper lipid is about 39-65 mol %, about 39-59 mol %, about 40-60 mol %, about 40-65 mol %, about 40-59 mol %, about 43-65 mol %, about 43-60 mol %, about 43-59 mol %, or about 50-65 mol %, about 50-59 mol %, about 59 mol %, or about 43.5 mol %. In certain embodiments, the amount of the PEG lipid is about 0.5-1.8 mol %, about 0.8-1.6 mol %, about 0.8-1.5 mol %, 0.9-1.8 mol %, about 0.9-1.6 mol %, about 0.9- 1.5 mol %, 1-1.8 mol %, about 1-1.6 mol %, about 1-1.5 mol %, about 1 mol %, or about 1.5 mol %. [00380] In some embodiments, the cargo includes an mRNA encoding an RNA-guided DNA-binding agent (e.g. a Cas nuclease, a Class 2 Cas nuclease, or Cas9), or a gRNA or a nucleic acid encoding a gRNA, or a combination of mRNA and gRNA. In one embodiment, a
Attorney Docket: 01155-0064-00PCT lipid nucleic acid assembly composition may comprise a Lipid A or its equivalents, or an amine lipid as provided in WO2020219876; or Lipid D or an amine lipid provided in WO2020/072605. In some aspects, the amine lipid is Lipid A, or Lipid D. In some aspects, the amine lipid is a Lipid A equivalent, e.g. an analog of Lipid A, or an amine lipid provided in WO2020/219876. In certain aspects, the amine lipid is an acetal analog of Lipid A, optionally, an amine lipid provided in WO2020/219876. In some aspects, the amine lipid is a Lipid D or an amine lipid found in in W2020072605. In various embodiments, a lipid nucleic acid assembly composition comprises an amine lipid, a neutral lipid, a helper lipid, and a PEG lipid. In some embodiments, the helper lipid is cholesterol. In some embodiments, the neutral lipid is DSPC. In specific embodiments, PEG lipid is PEG2k-DMG. In some embodiments, a lipid nucleic acid assembly composition may comprise a Lipid A, a helper lipid, a neutral lipid, and a PEG lipid. In some embodiments, a lipid nucleic acid assembly composition comprises an amine lipid, DSPC, cholesterol, and a PEG lipid. In some embodiments, the lipid nucleic acid assembly composition comprises a PEG lipid comprising DMG. In some embodiments, the amine lipid is selected from Lipid A, and an equivalent of Lipid A, including an acetal analog of Lipid A, or an amine lipid provided in WO2020/219876; or Lipid D or an amine lipid provided in WO2020/072605. In additional embodiments, a lipid nucleic acid assembly composition comprises Lipid A, cholesterol, DSPC, and PEG2k-DMG. In additional embodiments, a lipid nucleic acid assembly composition comprises Lipid D, cholesterol, DSPC, and PEG2k-DMG. [00381] Embodiments of the present disclosure also provide lipid compositions described according to the molar ratio between the positively charged amine groups of the amine lipid (N) and the negatively charged phosphate groups (P) of the nucleic acid to be encapsulated. This may be mathematically represented by the equation N/P. In some embodiments, a lipid nucleic acid assembly composition may comprise a lipid component that comprises an amine lipid, a helper lipid, a neutral lipid, and a helper lipid; and a nucleic acid component, wherein the N/P ratio is about 3 to 10. In some embodiments, the LNPs comprise molar ratios of an amine lipid to RNA/DNA phosphate (N:P) of about 4.5, 5.0, 5.5, 6.0, or 6.5. In some embodiments, a lipid nucleic acid assembly composition may comprise a lipid component that comprises an amine lipid, a helper lipid, a neutral lipid, and a helper lipid; and an RNA component, wherein the N/P ratio is about 3 to 10. In one embodiment, the N/P ratio may be about 5-7. In one embodiment, the N/P ratio may be about 4.5-8. In one embodiment, the N/P ratio may be about 6. In one embodiment, the N/P ratio may be 6 ±1. In one embodiment, the N/P ratio may be about 6 ± 0.5. In some embodiments, the N/P ratio will be ±30%, ±25%,
Attorney Docket: 01155-0064-00PCT ±20%, ±15%, ±10%, ±5%, or ±2.5% of the target N/P ratio. In some embodiments, lipid nucleic acid assembly inter-lot variability will be less than 15%, less than 10% or less than 5%. [00382] In some embodiments, the lipid nucleic acid assembly comprises an RNA component, which may comprise an mRNA, such as an mRNA encoding a Cas nuclease. In one embodiment, RNA component may comprise a Cas9 mRNA. In some compositions comprising an mRNA encoding a Cas nuclease, the lipid nucleic acid assembly further comprises a gRNA nucleic acid, such as a gRNA. In some embodiments, the RNA component comprises a Cas nuclease mRNA and a gRNA. In some embodiments, the RNA component comprises a Class 2 Cas nuclease mRNA and a gRNA. [00383] In some embodiments, a lipid nucleic acid assembly composition may comprise an mRNA encoding a Cas nuclease such as a Class 2 Cas nuclease, an amine lipid, a helper lipid, a neutral lipid, and a PEG lipid. In certain lipid nucleic acid assembly compositions comprising an mRNA encoding a Cas nuclease such as a Class 2 Cas nuclease, the helper lipid is cholesterol. In other compositions comprising an mRNA encoding a Cas nuclease such as a Class 2 Cas nuclease, the neutral lipid is DSPC. In additional embodiments comprising an mRNA encoding a Cas nuclease such as a Class 2 Cas nuclease, the PEG lipid is PEG2k-DMG or PEG2k-C11. In specific compositions comprising an mRNA encoding a Cas nuclease such as a Class 2 Cas nuclease, the amine lipid is selected from Lipid A and its equivalents, such as an acetal analog of Lipid A, or amine lipids provided in WO2020/219876; or Lipid D and amine lipids provided in WO2020/072605. [00384] In some embodiments, a lipid nucleic acid assembly composition may comprise a gRNA. In some embodiments, a lipid nucleic acid assembly composition may comprise an amine lipid, a gRNA, a helper lipid, a neutral lipid, and a PEG lipid. In certain lipid nucleic acid assembly compositions comprising a gRNA, the helper lipid is cholesterol. In some compositions comprising a gRNA, the neutral lipid is DSPC. In additional embodiments comprising a gRNA, the PEG lipid is PEG2k-DMG or PEG2k-C11. In some embodiments, the amine lipid is selected from Lipid A and its equivalents, such as an acetal analog of Lipid A, or amine lipids provided in WO2020/219876 and their equivalents; or Lipid D and amine lipids provided in WO2020/072605 and their equivalents. [00385] In one embodiment, a lipid nucleic acid assembly composition may comprise an sgRNA. In one embodiment, a lipid nucleic acid assembly composition may comprise a Cas9 sgRNA. In one embodiment, a lipid nucleic acid assembly composition may comprise a Cpf1 sgRNA. In some compositions comprising an sgRNA, the lipid nucleic acid assembly includes an amine lipid, a helper lipid, a neutral lipid, and a PEG lipid. In certain compositions
Attorney Docket: 01155-0064-00PCT comprising an sgRNA, the helper lipid is cholesterol. In other compositions comprising an sgRNA, the neutral lipid is DSPC. In additional embodiments comprising an sgRNA, the PEG lipid is PEG2k-DMG or PEG2k-C11. In some embodiments, the amine lipid is selected from Lipid A and its equivalents, such as acetal analogs of Lipid A, or amine lipids provided in WO2020/219876; or Lipid D and amine lipids provided in WO2020/072605. [00386] In some embodiments, a lipid nucleic acid assembly composition comprises an mRNA encoding a Cas nuclease and a gRNA, which may be an sgRNA. In one embodiment, a lipid nucleic acid assembly composition may comprise an amine lipid, an mRNA encoding a Cas nuclease, a gRNA, a helper lipid, a neutral lipid, and a PEG lipid. In certain compositions comprising an mRNA encoding a Cas nuclease and a gRNA, the helper lipid is cholesterol. In some compositions comprising an mRNA encoding a Cas nuclease and a gRNA, the neutral lipid is DSPC. In additional embodiments comprising an mRNA encoding a Cas nuclease and a gRNA, the PEG lipid is PEG2k-DMG or PEG2k-C11. In some embodiments, the amine lipid is selected from Lipid A and its equivalents, such as acetal analogs of Lipid A, or amine lipids provided in WO2020/219876; or Lipid D and amine lipids provided in WO2020/072605. [00387] In some embodiments, the lipid nucleic acid assembly compositions include a Cas nuclease mRNA, such as a Class 2 Cas mRNA and at least one gRNA. In some embodiments, the lipid nucleic acid assembly composition includes a ratio of gRNA to Cas nuclease mRNA, such as Class 2 Cas nuclease mRNA from about 25:1 to about 1:25 wt/wt. In some embodiments, the lipid nucleic acid assembly formulation includes a ratio of gRNA to Cas nuclease mRNA, such as Class 2 Cas nuclease mRNA from about 10:1 to about 1:10. In some embodiments, the lipid nucleic acid assembly formulation includes a ratio of gRNA to Cas nuclease mRNA, such as Class 2 Cas nuclease mRNA from about 8:1 to about 1:8. As measured herein, the ratios are by weight. In some embodiments, the lipid nucleic acid assembly formulation includes a ratio of gRNA to Cas nuclease mRNA, such as Class 2 Cas mRNA from about 5:1 to about 1:5. In some embodiments, ratio range is about 3:1 to 1:3, about 2:1 to 1:2, about 5:1 to 1:2, about 5:1 to 1:1, about 3:1 to 1:2, about 3:1 to 1:1, about 3:1, about 2:1 to 1:1. In some embodiments, the gRNA to mRNA ratio is about 3:1 or about 2:1. In some embodiments the ratio of gRNA to Cas nuclease mRNA, such as Class 2 Cas nuclease is about 1:1. In some embodiments the ratio of gRNA to Cas nuclease mRNA, such as Class 2 Cas nuclease is about 1:2. The ratio may be about 25:1, 10:1, 5:1, 3:1, 2:1, 1:1, 1:2, 1:3, 1:5, 1:10, or 1:25. [00388] The lipid nucleic acid assembly compositions disclosed herein may include a template nucleic acid. The template nucleic acid may be co-formulated with an mRNA
Attorney Docket: 01155-0064-00PCT encoding a Cas nuclease, such as a Class 2 Cas nuclease mRNA. In some embodiments, the template nucleic acid may be co-formulated with a guide RNA. In some embodiments, the template nucleic acid may be co-formulated with both an mRNA encoding a Cas nuclease and a guide RNA. In some embodiments, the template nucleic acid may be formulated separately from an mRNA encoding a Cas nuclease or a guide RNA. The template nucleic acid may be delivered with, or separately from the lipid nucleic acid assembly compositions. In some embodiments, the template nucleic acid may be single- or double-stranded, depending on the desired repair mechanism. The template may have regions of homology to the target DNA, or to sequences adjacent to the target DNA. [00389] In some embodiments, a lipid nucleic acid assemblies are formed by mixing an aqueous RNA solution with an organic solvent-based lipid solution, e.g., 100% ethanol. Suitable solutions or solvents include or may contain: water, PBS, Tris buffer, NaCl, citrate buffer, ethanol, chloroform, diethylether, cyclohexane, tetrahydrofuran, methanol, isopropanol. A pharmaceutically acceptable buffer, e.g., for in vivo administration of lipid nucleic acid assemblies, may be used. In some embodiments, a buffer is used to maintain the pH of the composition comprising lipid nucleic acid assemblies at or above pH 6.5. In some embodiments, a buffer is used to maintain the pH of the composition comprising lipid nucleic acid assemblies at or above pH 7.0. In some embodiments, the composition has a pH ranging from about 7.2 to about 7.7. In additional embodiments, the composition has a pH ranging from about 7.3 to about 7.7 or ranging from about 7.4 to about 7.6. In further embodiments, the composition has a pH of about 7.2, 7.3, 7.4, 7.5, 7.6, or 7.7. The pH of a composition may be measured with a micro pH probe. In some embodiments, a cryoprotectant is included in the composition. Non-limiting examples of cryoprotectants include sucrose, trehalose, glycerol, DMSO, and ethylene glycol. Exemplary compositions may include up to 10% cryoprotectant, such as, for example, sucrose. In some embodiments, the lipid nucleic acid assembly composition may include about 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10% cryoprotectant. In some embodiments, the lipid nucleic acid assembly composition may include about 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10% sucrose. In some embodiments, the lipid nucleic acid assembly composition may include a buffer. In some embodiments, the buffer may comprise a phosphate buffer (PBS), a Tris buffer, a citrate buffer, and mixtures thereof. In some exemplary embodiments, the buffer comprises NaCl. In some embodiments, NaCl is omitted. Exemplary amounts of NaCl may range from about 20 mM to about 45 mM. Exemplary amounts of NaCl may range from about 40 mM to about 50 mM. In some embodiments, the amount of NaCl is about 45 mM. In some embodiments, the buffer is a Tris buffer. Exemplary amounts of Tris may range
Attorney Docket: 01155-0064-00PCT from about 20 mM to about 60 mM. Exemplary amounts of Tris may range from about 40 mM to about 60 mM. In some embodiments, the amount of Tris is about 50 mM. In some embodiments, the buffer comprises NaCl and Tris. Certain exemplary embodiments of the lipid nucleic acid assembly compositions contain 5% sucrose and 45 mM NaCl in Tris buffer. In other exemplary embodiments, compositions contain sucrose in an amount of about 5% w/v, about 45 mM NaCl, and about 50 mM Tris at pH 7.5. The salt, buffer, and cryoprotectant amounts may be varied such that the osmolality of the overall formulation is maintained. For example, the final osmolality may be maintained at less than 450 mOsm/L. In further embodiments, the osmolality is between 350 and 250 mOsm/L. Certain embodiments have a final osmolality of 300 +/- 20 mOsm/L. [00390] In some embodiments, microfluidic mixing, T-mixing, or cross-mixing is used. In certain aspects, flow rates, junction size, junction geometry, junction shape, tube diameter, solutions, and/or RNA and lipid concentrations may be varied. Lipid nucleic acid assemblies or lipid nucleic acid assembly compositions may be concentrated or purified, e.g., via dialysis, tangential flow filtration, or chromatography. The lipid nucleic acid assemblies may be stored as a suspension, an emulsion, or a lyophilized powder, for example. In some embodiments, a lipid nucleic acid assembly composition is stored at 2-8° C, in certain aspects, the lipid nucleic acid assembly compositions are stored at room temperature. In additional embodiments, a lipid nucleic acid assembly composition is stored frozen, for example at -20° C or -80° C. In other embodiments, a lipid nucleic acid assembly composition is stored at a temperature ranging from about 0° C to about -80° C. Frozen lipid nucleic acid assembly compositions may be thawed before use, for example on ice, at 4° C, at room temperature, or at 25° C. Frozen lipid nucleic acid assembly compositions may be maintained at various temperatures, for example on ice, at 4° C, at room temperature, at 25° C, or at 37° C. [00391] In some embodiments, the concentration of the LNPs in the LNP composition is about 1-10 ug/mL, about 2-10 ug/mL, about 2.5-10 ug/mL, about 1-5 ug/mL, about 2-5 ug/mL, about 2.5-5 ug/mL, about 0.04 ug/mL, about 0.08 ug/mL, about 0.16 ug/mL, about 0.25 ug/mL, about 0.63 ug/mL, about 1.25 ug/mL, about 2.5 ug/mL, or about 5 ug/mL. [00392] In some embodiments, the lipid nucleic acid assembly composition comprises a stealth lipid, optionally wherein: (i) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50-60 mol % amine lipid such as Lipid A or Lipid D, about 8-10 mol % neutral lipid; and about 2.5-4 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic
Attorney Docket: 01155-0064-00PCT acid assembly composition is about 6; (ii) the lipid nucleic acid assembly composition comprises about 50-60 mol % amine lipid such as Lipid A or Lipid D; about 27-39.5 mol % helper lipid; about 8-10 mol % neutral lipid; and about 2.5-4 mol % stealth lipid (e.g., a PEG lipid), wherein the N/P ratio of the lipid nucleic acid assembly composition is about 5-7 (e.g., about 6); (iii) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50-60 mol % amine lipid such as Lipid A or Lipid D; about 5-15 mol % neutral lipid; and about 2.5-4 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic acid assembly composition is about 3-10; (iv) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 40-60 mol % amine lipid such as Lipid A or Lipid D; about 5-15 mol % neutral lipid; and about 2.5-4 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic acid assembly composition is about 6; (v) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50-60 mol % amine lipid such as Lipid A or Lipid D; about 5-15 mol % neutral lipid; and about 1.5-10 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic acid assembly composition is about 6; (vi) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 40-60 mol % amine lipid such as Lipid A or Lipid D; about 0-10 mol % neutral lipid; and about 1.5-10 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic acid assembly composition is about 3-10; (vii) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 40-60 mol % amine lipid such as Lipid A or Lipid D; less than about 1 mol % neutral lipid; and about 1.5-10 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic acid assembly composition is about 3-10; (viii) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 40-60 mol % amine lipid such as Lipid A or Lipid D; and about 1.5-10 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, wherein the N/P ratio of the LNP composition is about 3-10, and wherein the lipid
Attorney Docket: 01155-0064-00PCT nucleic acid assembly composition is essentially free of or free of neutral phospholipid; or (ix) the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50-60 mol % amine lipid such as Lipid A or Lipid D; about 8-10 mol-% neutral lipid; and about 2.5-4 mol % stealth lipid (e.g., a PEG lipid), wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the lipid nucleic acid assembly composition is about 3-7. [00393] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50 mol % amine lipid such as Lipid A or Lipid D; about 9 mol % neutral lipid such as DSPC; about 3 mol % of stealth lipid such as a PEG lipid, such as PEG2k-DMG, and the remainder of the lipid component is helper lipid such as cholesterol wherein the N/P ratio of the lipid nucleic acid assembly composition is about 6. [00394] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50 mol % Lipid A; about 9 mol % DSPC; about 3 mol % of PEG2k-DMG, and the remainder of the lipid component is cholesterol wherein the N/P ratio of the lipid nucleic acid assembly composition is about 6. [00395] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 50 mol % Lipid A; about 9 mol % DSPC; about 1.5 mol % of PEG2k-DMG, and the remainder of the lipid component (39.5 mol %) is cholesterol wherein the N/P ratio of the lipid nucleic acid assembly composition is about 6. [00396] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 35 mol % Lipid A; about 15 mol % DSPC; about 2.5 mol % of PEG2k-DMG, and the remainder of the lipid component (47.5 mol %) is cholesterol wherein the N/P ratio of the lipid nucleic acid assembly composition is about 6. [00397] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 35 mol % Lipid A; about 15 mol % neutral lipid; about 47.5 mol % helper lipid; and about 2.5 mol % stealth lipid (e.g., PEG lipid), and wherein the N/P ratio of the LNP composition is about 3-7. [00398] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 35 mol % Lipid D; about 15 mol % neutral lipid; about 47.5 mol % helper lipid; and about 2.5 mol % stealth lipid (e.g., PEG lipid), and wherein the N/P ratio of the LNP composition is about 3-7.
Attorney Docket: 01155-0064-00PCT [00399] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 25-45 mol % amine lipid, such as Lipid A; about 10-30 mol % neutral lipid; about 25-65 mol % helper lipid; and about 1.5-3.5 mol % stealth lipid (e.g., PEG lipid), and wherein the N/P ratio of the LNP composition is about 3-7. [00400] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component, wherein: a. the amount of the amine lipid is about 29-44 mol % of the lipid component; the amount of the neutral lipid is about 11-28 mol % of the lipid component; the amount of the helper lipid is about 28-55 mol % of the lipid component; and the amount of the PEG lipid is about 2.3-3.5 mol % of the lipid component b. the amount of the amine lipid is about 29-38 mol % of the lipid component; the amount of the neutral lipid is about 11-20 mol % of the lipid component; the amount of the helper lipid is about 43-55 mol % of the lipid component; and the amount of the PEG lipid is about 2.3-2.7 mol % of the lipid component; c. the amount of the amine lipid is about 25-34 mol % of the lipid component; the amount of the neutral lipid is about 10-20 mol % of the lipid component; the amount of the helper lipid is about 45-65 mol % of the lipid component; and the amount of the PEG lipid is about 2.5-3.5 mol % of the lipid component; or d. the amount of the amine lipid is about 30-43 mol % of the lipid component; the amount of the neutral lipid is about 10-17 mol % of the lipid component; the amount of the helper lipid is about 43.5-56 mol % of the lipid component; and the amount of the PEG lipid is about 1.5-3 mol % of the lipid component. [00401] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component and the lipid component comprises: about 25-50 mol % amine lipid, such as Lipid D; about 7-25 mol % neutral lipid; about 39-65 mol % helper lipid; and about 0.5-1.8 mol % stealth lipid (e.g., PEG lipid), and wherein the N/P ratio of the LNP composition is about 3-7. [00402] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component wherein the amount of the amine lipid is about 30-45 mol % of the lipid component; or about 30-40 mol % of the lipid component; optionally about 30 mol %, 40 mol %, or 50 mol % of the lipid component. In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component wherein the amount of the neutral lipid is about 10-
Attorney Docket: 01155-0064-00PCT 20 mol % of the lipid component; or about 10-15 mol % of the lipid component; optionally about 10 mol % or 15 mol % of the lipid component. In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component wherein the amount of the helper lipid is about 50-60 mol % of the lipid component; about 39-59 mol % of the lipid component; or about 43.5-59 mol % of the lipid component; optionally about 59 mol % of the lipid component; about 43.5 mol % of the lipid component; or about 39 mol % of the lipid component. In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component wherein the amount of the PEG lipid is about 0.9-1.6 mol % of the lipid component; or about 1-1.5 mol % of the lipid component; optionally about 1 mol % of the lipid component or about 1.5 mol % of the lipid component [00403] In some embodiments, the lipid nucleic acid assembly composition comprises a lipid component, wherein: e. the amount of the ionizable lipid is about 27-40 mol % of the lipid component; the amount of the neutral lipid is about 10-20 mol % of the lipid component; the amount of the helper lipid is about 50-60 mol % of the lipid component; and the amount of the PEG lipid is about 0.9-1.6 mol % of the lipid component; f. the amount of the ionizable lipid is from about 30-45 mol % of the lipid component; the amount of the neutral lipid is from about 10-15 mol % of the lipid component; the amount of the helper lipid is from about 39-59 mol % of the lipid component; and the amount of the PEG lipid is from about 1-1.5 mol % of the lipid component; g. the amount of the ionizable lipid is about 30 mol % of the lipid component; the amount of the neutral lipid is about 10 mol % of the lipid component; the amount of the helper lipid is about 59 mol % of the lipid component; and the amount of the PEG lipid is about 1-1.5 mol % of the lipid component; h. the amount of the ionizable lipid is about 40 mol % of the lipid component; the amount of the neutral lipid is about 15 mol % of the lipid component; the amount of the helper lipid is about 43.5 mol % of the lipid component; and the amount of the PEG lipid is about 1.5 mol % of the lipid component; or i. the amount of the ionizable lipid is about 50 mol % of the lipid component; the amount of the neutral lipid is about 10 mol % of the lipid component; the amount of the helper lipid is about 39 mol % of the lipid component; and the amount of the PEG lipid is about 1 mol % of the lipid component.
Attorney Docket: 01155-0064-00PCT [00404] In some embodiments, the LNP has a diameter of about 1-250 nm, 10-200 nm, about 20-150 nm, about 50-150 nm, about 50-100 nm, about 50-120 nm, about 60-100 nm, about 75- 150 nm, about 75-120 nm, or about 75-100 nm. In some embodiments, the LNP has a diameter of less than 100 nm. In some embodiments, the LNP composition comprises a population of the LNP with an average diameter of about 10-200 nm, about 20-150 nm, about 50-150 nm, about 50-100 nm, about 50-120 nm, about 60-100 nm, about 75-150 nm, about 75-120 nm, or about 75-100 nm. In some embodiments, the LNP has an average diameter of less than 100 nm. [00405] In some embodiments, the lipid nucleic acid assembly composition comprises: about 40-60 mol-% amine lipid; about 5-15 mol-% neutral lipid; and about 1.5-10 mol-% PEG lipid, wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the LNP composition is about 3-10. In some embodiments, the lipid nucleic acid assembly composition comprises: about 50-60 mol-% amine lipid; about 8-10 mol-% neutral lipid; and about 2.5-4 mol-% PEG lipid, wherein the remainder of the lipid component is helper lipid, and wherein the N/P ratio of the LNP composition is about 3-8. In some embodiments, the lipid nucleic acid assembly composition comprises: about 50-60 mol-% amine lipid; about 5-15 mol- % DSPC; and about 2.5-4 mol-% PEG lipid, wherein the remainder of the lipid component is cholesterol, and wherein the N/P ratio of the LNP composition is 3-8 ±0.2. [00406] In embodiments, the average diameter is a Z-average diameter. In certain embodiments, the Z-average diameter is measured by dynamic light scattering (DLS) using methods known in the art. For example, average particle size and polydispersity can be measured by dynamic light scattering (DLS) using a Malvern Zetasizer DLS instrument. LNP samples are diluted with PBS buffer prior to being measured by DLS. Z-average diameter and number average diameter along with a polydispersity index (pdi) can be determined. The Z average is the intensity weighted mean hydrodynamic size of the ensemble collection of particles. The number average is the particle number weighted mean hydrodynamic size of the ensemble collection of particles. A Malvern Zetasizer instrument can also be used to measure the zeta potential of the LNP using methods known in the art. D. DNA-Dependent Protein Kinase Inhibitors [00407] In some embodiments, the cells are contacted with a DNA-dependent protein kinase (DNA-PK), which is a nuclear serine/threonine kinase that has been shown to be essential in DNA double stranded break repair machinery. In mammals, the predominant pathway for repair of double stranded DNA breaks is the non-homologous end joining (NHEJ) pathway
Attorney Docket: 01155-0064-00PCT which is functional regardless of the phase of the cell cycle and acts by removing non-ligatable ends and ligating ends of double strand breaks. DNA-PK inhibitors (DNA-PKi) are a structurally diverse class of inhibitors of DNA-PK, and the NHEJ pathway. Exemplary DNA- PKi are provided, for example, in WO03024949, WO2014159690A1, and WO2018114999. [00408] DNA-dependent protein kinase (DNA-PK) is a nuclear serine/threonine kinase that has been shown to be essential in DNA double stranded break repair machinery. In mammals, the predominant pathway for repair of double stranded DNA breaks is the non-homologous end joining (NHEJ) pathway which is functional regardless of the phase of the cell cycle and acts by removing non-ligatable ends and ligating ends of double strand breaks. DNA-PK inhibitors (DNA-PKi) are a structurally diverse class of inhibitors of DNA-PK, and the NHEJ pathway. Exemplary DNA-PKi are provided, for example, in WO03024949, WO2014159690A1, and WO2018114999. [00409] In preferred embodiments, the disclosure relates to a DNAPKI Compound 1 that is .

[00410] In preferred to a DNAPKI Compound 3 that is . [00411] In preferred
a DNAPKI Compound 4 that is
Attorney Docket: 01155-0064-00PCT . [00412] In certain of the compositions described
herein, wherein the is about 1 µM or less, for example, about 0.25 µM or less, such as about 0.1-1 µM, preferably about 0.1-0.5 µM. [00413] In some embodiments, the DNAPKI is formed according to the methods set forth in WO2018114999, which is incorporated by reference. [00414] Exemplary DNA-PKi include, but are not limited to, Compound 1, Compound 3 and Compound 4. In some embodiments, the DNAPKi is Compound 1. In some embodiments, the DNAPKI is Compound 3. In some embodiments, the DNAPKi is Compound 4. 1. Synthesis of DNA-Dependent Protein Kinase Inhibitors a) Compound 1 [00415] Intermediate 1a: (E)-N,N-dimethyl-N'-(4-methyl-5-nitropyridin-2- yl)formimidamide [00416] To a solution of 4-

(5 g, 1.0 equiv.) in toluene (0.3 M) was added DMF-DMA (3.0 equiv.). The mixture was stirred at 110 °C for 2 h. The reaction mixture was concentrated under reduced pressure to give a residue and purified by column chromatography to afford product as a yellow solid (59%).1H NMR (400 MHz, (CD3)2SO) δ 8.82 (s, 1H), 8.63 (s, 1H), 6.74 (s, 1H), 3.21 (m, 6H). [00417] Intermediate 1b: (E)-N-hydroxy-N'-(4-methyl-5-nitropyridin-2-yl)formimidamide [00418] To a solution of
in MeOH (0.2 M) was added NH2OH ^HCl (2.0 equiv.). The reaction mixture was stirred at 80 °C for 1 h. The reaction mixture was filtered, and the filtrate was concentrated under reduced pressure to give a residue.
Attorney Docket: 01155-0064-00PCT The residue was partitioned between H2O and EtOAc, followed by 2x extraction with EtOAc. The organic phases were concentrated under reduced pressure to give a residue and purified by column chromatography to afford product as a white solid (66%). 1H NMR (400 MHz, (CD3)2SO) δ 10.52 (d, J = 3.8 Hz, 1H), 10.08 (dd, J = 9.9, 3.7 Hz, 1H), 8.84 (d, J = 3.8 Hz, 1H), 7.85 (dd, J = 9.7, 3.8 Hz, 1H), 7.01 (d, J = 3.9 Hz, 1H), 3.36 (s, 3 H). [00419] Intermediate 1c: 7-methyl-6-nitro-[1,2,4]triazolo[1,5-a]pyridine [00420] To a solution of 1.0 equiv.) in THF (0.4 M) was added

trifluoroacetic anhydride (1.0 equiv.) was stirred at 25 °C for 18 h. The reaction mixture was filtered, and the filtrate was concentrated under reduced pressure to give a residue. The residue was purified by column chromatography to afford product as a white solid (44%).1H NMR (400 MHz, CDCl3) δ 9.53 (s, 1H), 8.49 (s, 1H), 7.69 (s, 1H), 2.78 (d, J = 1.0 Hz, 3H). [00421] Intermediate 1d: 7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-amine [00422] To a mixture of Pd/C
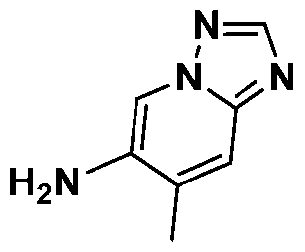
equiv.) in EtOH (0.1 M) was added Intermediate 1c (1.0 equiv. and ammonium formate (5.0 equiv.). The mixture was heated at 105 °C for 2 h. The reaction mixture was filtered, and the filtrate was concentrated under reduced pressure to give a residue. The residue was purified by column chromatography to afford product as a pale brown solid.1H NMR (400 MHz, (CD3)2SO) δ 8.41 (s, 2H), 8.07 (d, J = 9.0 Hz, 2H), 7.43 (s, 1H), 2.22 (s, 3H). [00423] Intermediate 1e: ethyl 2-chloro-4-((tetrahydro-2H-pyran-4-yl)amino)pyrimidine-5- carboxylate

Attorney Docket: 01155-0064-00PCT [00424] To a solution of tetrahydropyran-4-amine (5 g, 1.0 equiv.) and ethyl 2,4- dichloropyrimidine-5-carboxylate (1.0 equiv.) in MeCN (0.25 – 2.0 M) was added K2CO3 (1.0 –3.0 equiv.). The mixture was stirred at 20-25 °C for at least 12 h. The reaction mixture was filtered, and the filtrate was concentrated under reduced pressure to give a residue. The residue was purified by column chromatography to afford product as a pale yellow solid (21%). 1H NMR (400 MHz, (CD3)2SO) δ 8.60 (s, 1H), 8.29 (d, J = 7.7 Hz, 1H), 4.28 (q, J = 7.1 Hz, 2H), 4.14 (dtt, J = 11.3, 8.3, 4.0 Hz, 1H), 3.82 (dt, J = 12.1, 3.6 Hz, 2H), 3.57 (s, 1H), 1.87 – 1.78 (m, 2H), 1.76 – 1.67 (m, 1H), 1.54 (qd, J = 10.9, 4.3 Hz, 2H), 1.28 (t, J = 7.1 Hz, 3H). [00425] Intermediate 1f: 2-chloro-4-((tetrahydro-2H-pyran-4-yl)amino)pyrimidine-5- carboxylic acid [00426] To a solution of LiOH THF/H2O (0.25 – 1.0 M) was added
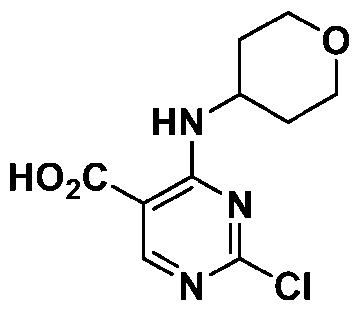
Intermediate 1e (3.0 g, 1.0 equiv.). The mixture was stirred at 25 °C for 12 h. The mixture was concentrated under reduced pressure to remove THF. The residue was adjusted pH to 2 by 2 M HCl, and the resulting precipitate was collected by filtration, washed with water, and dried under vacuum to get a residue. The residue was purified by column chromatography to afford product as a white solid (74%) or used directly as a crude product. [00427] Intermediate 1g: 2-chloro-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8- one [00428] To a solution of

equiv.) in MeCN (0.2 – 0.5 M) was added Et3N (1.0 equiv.). The mixture was stirred at 25 °C for 30 min. Then DPPA (1.0 equiv.) was added to the mixture. The mixture was stirred at 100 °C for at least 7 h. The reaction mixture was poured into water, and the resulting precipitate was collected by filtration, washed with water, and dried under vacuum to get a residue. The residue was purified by column chromatography to afford product as a white solid (56%).1H NMR (400 MHz, CDCl3) δ 9.50
Attorney Docket: 01155-0064-00PCT (s, 1H), 8.09 (s, 1H), 4.53 (tt, J = 12.4, 4.2 Hz, 1H), 4.07 (dt, J = 9.5, 4.8 Hz, 2H), 3.48 (td, J = 12.1, 1.9 Hz, 2H), 2.69 (qd, J = 12.5, 4.7 Hz, 2H), 1.67 (dd, J = 12.1, 3.9 Hz, 2H). [00429] Intermediate 1h: 2-chloro-7-methyl-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H- purin-8-one [00430] To a mixture of 1.0 equiv.) and NaOH (5.0 equiv.) in 1:1
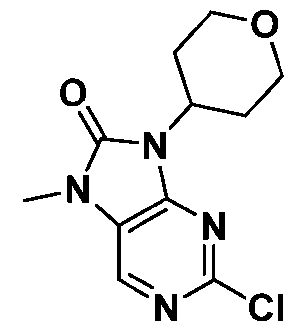
THF/H2O (0.25-1.0 M) was added equiv.). The reaction mixture was stirred at 25 °C for 12 h. The reaction mixture was concentrated under reduced pressure to give a residue and purified by column chromatography to afford product as a white solid (47%).1H NMR (400 MHz, (CD3)2SO) δ 8.34 (s, 1H), 4.43 (ddt, J = 12.2, 8.5, 4.2 Hz, 1H), 3.95 (dd, J = 11.5, 4.6 Hz, 2H), 3.43 (td, J = 12.1, 1.9 Hz, 2H), 2.45 (s, 3H), 2.40 (td, J = 12.5, 4.7 Hz, 2H), 1.66 (ddd, J = 12.2, 4.4, 1.9 Hz, 2H). [00431] Compound 1: 7-methyl-2-((7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino)-9- (tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (Compound 1) [00432] A mixture of
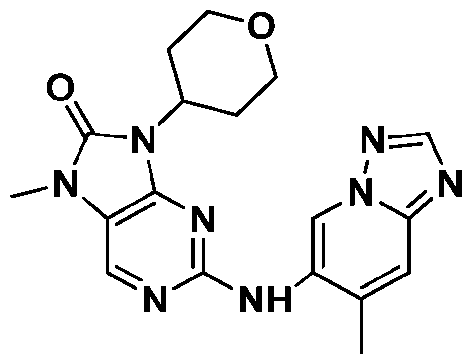
, Intermediate 1d (1.0 equiv.), Pd(dppf)Cl2 (0.1 - 0.2 equiv.), XantPhos (0.1 - 0.2 equiv.) and Cs2CO3 (2.0 equiv.) in DMF (0.05 – 0.3 M) was degassed and purged 3x with N2 and the mixture was stirred at 100-130 °C for at least 12 h under N2 atmosphere. The reaction mixture was then poured into water and extracted 3x with DCM. The combined organic phase was washed with brine, dried with anhydrous Na2SO4, filtered, and the filtrate was concentrated in vacuum. The residue was purified by column chromatography to afford product as a pale yellow solid. 1H NMR (400 MHz, (CD3)2SO) δ 9.13 (s, 1H), 8.69 (s, 1H), 8.39 (s, 1H), 8.10 (s, 1H), 7.72 (s, 1H), 4.50 – 4.36 (m, 1H), 3.98 (dd, J = 11.6, 4.4 Hz, 2H), 3.44 (d, J = 11.9 Hz, 2H), 3.32 (s, 3H), 2.44 – 2.38 (m, 3H), 1.69 (d, J = 11.6 Hz, 2H). MS: 381.3 m/z [M+H].
Attorney Docket: 01155-0064-00PCT b) Compound 3 [00433] Intermediate 3a: ethyl 2-chloro-4-((4,4-difluorocyclohexyl)amino)pyrimidine-5- carboxylate [00434] Intermediate 3a was 2,4-dichloropyrimidine-5-carboxylate

and 4,4-difluorocyclohexanamine the method employed in Intermediate 1e. 1H NMR (400 MHz, (CD3)2SO) δ 8.61 (s, 1H), 8.30 (d, J = 7.7 Hz, 1H), 4.29 (q, J = 7.1 Hz, 2H), 4.19 – 4.09 (m, 1H), 2.09 – 1.90 (m, 6H), 1.69 – 1.58 (m, 2H), 1.29 (t, J = 7.1 Hz, 3H). [00435] Intermediate 3b: 2-chloro-4-((4,4-difluorocyclohexyl)amino)pyrimidine-5- carboxylic acid [00436] Intermediate 3b was
Intermediate 3a using the method employed in Intermediate 1f. 1H NMR (400 MHz, (CD3)2SO) δ 13.77 (s, 1H), 8.57 (s, 1H), 8.53 (d, J = 7.8 Hz, 1H), 4.12 (d, J = 10.2 Hz, 1H), 2.14 – 1.89 (m, 6H), 1.62 (ddt, J = 17.0, 10.3, 6.0 Hz, 2H). [00437] Intermediate 3c: 2-chloro-9-(4,4-difluorocyclohexyl)-7,9-dihydro-8H-purin-8-one [00438] Intermediate 3c was
from Intermediate 3b using the method employed in Intermediate 1g.1H NMR (400 MHz, (CD3)2SO) δ 11.76 – 11.65 (m, 1H), 8.20 (s, 1H), 4.47 (dq, J = 12.6, 6.2, 4.3 Hz, 1H), 2.34 – 1.97 (m, 6H), 1.90 (d, J = 12.9 Hz, 2H).
Attorney Docket: 01155-0064-00PCT [00439] Intermediate 3d: 2-chloro-9-(4,4-difluorocyclohexyl)-7-methyl-7,9-dihydro-8H- purin-8-one [00440] To a mixture of 1.0 equiv.), NaOH (5.0 equiv.) in 5:1
THF/H2O (0.3 M) was added MeI . was stirred at 20 °C for 12 h under N2 atmosphere. The reaction mixture was concentrated under reduced pressure to give a residue and purified by column chromatography to afford product as a yellow solid (47%).1H NMR (400 MHz, CDCl3) δ 8.01 (s, 1H), 4.53 – 4.39 (m, 1H), 3.43 (s, 3H), 2.73 (qd, J = 12.7, 12.1, 3.8 Hz, 2H), 2.32 – 2.20 (m, 2H), 2.03 – 1.82 (m, 4H). [00441] Compound 3: 9-(4,4-difluorocyclohexyl)-7-methyl-2-((7-methyl- [1,2,4]triazolo[1,5-a]pyridin-6-yl)amino)-7,9-dihydro-8H-purin-8-one (Compound 3) [00442] Compound 3 was

1d and Intermediate 3d using the method employed for Compound 1, followed by a purification by reverse-phase HPLC. 1H NMR (400 MHz, (CD3)2SO) δ 9.03 (s, 1H), 8.66 (s, 1H), 8.38 (s, 1H), 8.10 (s, 1H), 7.71 (d, J = 1.4 Hz, 1H), 4.36 (d, J = 12.3 Hz, 1H), 3.31 (s, 3H), 2.38 (d, J = 1.0 Hz, 3H), 2.11 – 1.96 (m, 4H), 1.81 (d, J = 12.6 Hz, 2H). MS: 415.5 m/z [M+H]. c) Compound 4 [00443] Intermediate 4a: 8-methylene-1,4-dioxaspiro[4.5]decane [00444] To a solution of methyl

bromide (1.15 equiv.) in THF (0.6 M) was added n-BuLi (1.1 equiv.) at -78 °C dropwise, and the mixture was stirred at 0 °C for
Attorney Docket: 01155-0064-00PCT 1 h. Then, 1,4-dioxaspiro[4.5]decan-8-one (50 g, 1.0 equiv.) was added to the reaction mixture. The mixture was stirred at 25 °C for 12 h. The reaction mixture was poured into aq. NH4Cl at 0°C, diluted with H2O, and extracted 3x with EtOAc. The combined organic layers were concentrated under reduced pressure to give a residue and purified by column chromatography to afford product as a colorless oil (51%).1H NMR (400 MHz, CDCl3) δ 4.67 (s, 1H), 3.96 (s, 4 H), 2.82 (t, J = 6.4 Hz, 4 H), 1.70 (t, J = 6.4 Hz, 4 H). [00445] Intermediate 4b: 7,10-dioxadispiro[2.2.46.23]dodecane [00446] To a solution of equiv.) in toluene (3 M) was added ZnEt2

(2.57 equiv.) dropwise at -40 °C was stirred at -40 °C for 1 h. Then diiodomethane (6.0 equiv.) was added dropwise to the mixture at -40 °C under N2. The mixture was then stirred at 20 °C for 17 h under N2 atmosphere. The reaction mixture was poured into aq. NH4Cl at 0 °C and extracted 2x with EtOAc. The combined organic phases were washed with brine (20 mL), dried with anhydrous Na2SO4, filtered, and the filtrate was concentrated in vacuum. The residue was purified by column chromatography to afford product as a pale- yellow oil (73%). [00447] Intermediate 4c: spiro[2.5]octan-6-one [00448] To a solution of

1.0 equiv.) in 1:1 THF/H2O (1.0 M) was added TFA (3.0 equiv.). The mixture was stirred at 20 °C for 2 h under N2 atmosphere. The reaction mixture was concentrated under reduced pressure to remove THF, and the residue adjusted pH to 7 with 2 M NaOH (aq.). The mixture was poured into water and 3x extracted with EtOAc. The combined organic phase was washed with brine, dried with anhydrous Na2SO4, filtered, and the filtrate was concentrated in vacuum. The residue was purified by column chromatography to afford product as a pale-yellow oil (68%). 1H NMR (400 MHz, CDCl3) δ 2.35 (t, J = 6.6 Hz, 4H), 1.62 (t, J = 6.6 Hz, 4H), 0.42 (s, 4H). [00449] Intermediate 4d: N-(4-methoxybenzyl)spiro[2.5]octan-6-amine
Attorney Docket: 01155-0064-00PCT [00450] To a mixture of Intermediate 4c (2 g, 1.0 equiv.) and (4- methoxyphenyl)methanamine (1.1 equiv.) in DCM (0.3 M) was added AcOH (1.3 equiv.). The mixture was stirred at 20 °C for 1 h under N2 atmosphere. Then, NaBH(OAc)3 (3.3 equiv.) was added to the mixture at 0 °C, and the mixture was stirred at 20 °C for 17 h under N2 atmosphere. The reaction mixture was concentrated under reduced pressure to remove DCM, and the resulting residue was diluted with H2O and extracted 3x with EtOAc. The combined organic layers were washed with brine, dried over Na2SO4, filtered, and the filtrate was concentrated under reduced pressure to give a residue. The residue was purified by column chromatography to afford product as a gray solid (51%). 1H NMR (400 MHz, (CD3)2SO) δ 7.15 – 7.07 (m, 2H), 6.77 – 6.68 (m, 2H), 3.58 (s, 3H), 3.54 (s, 2H), 2.30 (ddt, J = 10.1, 7.3, 3.7 Hz, 1H), 1.69 – 1.62 (m, 2H), 1.37 (td, J = 12.6, 3.5 Hz, 2H), 1.12 – 1.02 (m, 2H), 0.87 – 0.78 (m, 2H), 0.13 – 0.04 (m, 2H). [00451] Intermediate 4e: spiro[2.5]octan-6-amine [00452] To a suspension of Pd/C equiv.) in MeOH (0.25 M) was added
Intermediate 4d (2 g, 1.0 equiv.) and the mixture was stirred at 80 °C at 50 Psi for 24 h under H2 atmosphere. The reaction mixture was filtered, and the filtrate was concentrated under reduced pressure to give a residue that was purified by column chromatography to afford product as a white solid. 1H NMR (400 MHz, (CD3)2SO) δ 2.61 (tt, J = 10.8, 3.9 Hz, 1H), 1.63 (ddd, J = 9.6, 5.1, 2.2 Hz, 2H), 1.47 (td, J = 12.8, 3.5 Hz, 2H), 1.21 – 1.06 (m, 2H), 0.82 – 0.72 (m, 2H), 0.14 – 0.05 (m, 2H). [00453] Intermediate 4f: ethyl 2-chloro-4-(spiro[2.5]octan-6-ylamino)pyrimidine-5- carboxylate [00454] Intermediate 4f was

Intermediate 4e using the method employed in Intermediate 1e.1H NMR (400 MHz, (CD3)2SO) δ 8.64 (s, 1H), 8.41 (d, J = 7.9 Hz, 1H), 4.33 (q, J = 7.1 Hz, 2H), 4.08 (d, J = 9.8 Hz, 1H), 1.90 (dd, J = 12.7, 4.8 Hz, 2H), 1.64 (t, J = 12.3 Hz, 2H), 1.52 (q, J = 10.7, 9.1 Hz, 2H), 1.33 (t, J = 7.1 Hz, 3H), 1.12 (d, J = 13.0 Hz, 2H), 0.40 – 0.21 (m, 4H).
Attorney Docket: 01155-0064-00PCT [00455] Intermediate 4g: 2-chloro-4-(spiro[2.5]octan-6-ylamino)pyrimidine-5-carboxylic acid [00456] Intermediate 4g was Intermediate 4f using the method
employed in Intermediate 1f.1H 2SO) δ 13.54 (s, 1H), 8.38 (d, J = 8.0 Hz, 1H), 8.35 (s, 1H), 3.82 (qt, J = 8.2, 3.7 Hz, 1H), 1.66 (dq, J = 12.8, 4.1 Hz, 2H), 1.47 – 1.34 (m, 2H), 1.33 – 1.20 (m, 2H), 0.86 (dt, J = 13.6, 4.2 Hz, 2H), 0.08 (dd, J = 8.3, 4.8 Hz, 4H). [00457] Intermediate 4h: 2-chloro-9-(spiro[2.5]octan-6-yl)-7,9-dihydro-8H-purin-8-one [00458] Intermediate 4h was
from Intermediate 4g using the method employed in Intermediate 1g. 1H NMR (400 MHz, (CD3)2SO) δ 11.68 (s, 1H), 8.18 (s, 1H), 4.26 (ddt, J = 12.3, 7.5, 3.7 Hz, 1H), 2.42 (qd, J = 12.6, 3.7 Hz, 2H), 1.95 (td, J = 13.3, 3.5 Hz, 2H), 1.82 – 1.69 (m, 2H), 1.08 – 0.95 (m, 2H), 0.39 (tdq, J = 11.6, 8.7, 4.2, 3.5 Hz, 4H). [00459] Intermediate 4i: 2-chloro-7-methyl-9-(spiro[2.5]octan-6-yl)-7,9-dihydro-8H-purin- 8-one [00460] Intermediate 4i was
from Intermediate 4h using the method employed in Intermediate 1h. 1H NMR (400 MHz, CDCl3) δ 7.57 (s, 1H), 4.03 (tt, J = 12.5, 3.9 Hz, 1H), 3.03 (s, 3H), 2.17 (qd, J = 12.6, 3.8 Hz, 2H), 1.60 (td, J = 13.4, 3.6 Hz, 2H), 1.47 – 1.34 (m, 2H), 1.07 (s, 1H), 0.63 (dp, J = 14.0, 2.5 Hz, 2H), -0.05 (s, 4H). [00461] Compound 4: 7-methyl-2-((7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino)-9- (spiro[2.5]octan-6-yl)-7,9-dihydro-8H-purin-8-one (Compound 4)
Attorney Docket: 01155-0064-00PCT [00462] Compound 4 was 4i and Intermediate 1d using the method employed in Compound

(CD3)2SO) δ 9.09 (s, 1H), 8.73 (s, 1H), 8.44 (s, 1H), 8.16 (s, 1H), 7.78 (s, 1H), 4.21 (t, J = 12.5 Hz, 1H), 3.36 (s, 3H), 2.43 (s, 3H), 2.34 (dt, J = 13.0, 6.5 Hz, 2H), 1.93 – 1.77 (m, 2H), 1.77 – 1.62 (m, 2H), 0.91 (d, J = 13.2 Hz, 2H), 0.31 (t, J = 7.1 Hz, 2H). MS: 405.5 m/z [M+H]. VII. Examples The following examples are provided to illustrate certain disclosed embodiments and are not to be construed as limiting the scope of this disclosure in any way. Example 1. Materials and Methods 1.1 In Vitro Transcription ("IVT") of mRNA [00463] Capped and polyadenylated mRNA containing N1-methyl pseudo-U was generated by in vitro transcription using a linearized plasmid DNA template and T7 RNA polymerase. The linearized plasmid DNA containing a T7 promoter and a sequence for transcription was linearized by restriction endonuclease digestion followed by heat inactivation of the reaction mixture and purified from enzyme and buffer salts. Messenger RNA was synthesized and purified using standard techniques known in the art. [00464] Messenger RNA was generated from plasmid DNA encoding an open reading frame. When the sequences cited in this paragraph are referred to below with respect to RNAs, it is understood that Ts should be replaced with Us (which can be modified nucleosides as described above). Messenger RNAs used in the Examples include a 5' cap and a 3' polyadenylation sequence e.g., up to 100 nts. Guide RNAs were chemically synthesized by commercial vendors or using standard in vitro synthesis techniques with modified nucleotides. 1.2 Lipid Nanoparticle Formulation [00465] In general, the lipid nanoparticle (LNP) components were dissolved in 100% ethanol at various molar ratios. The RNA cargos (e.g., Cas9 mRNA and sgRNA) were dissolved in 25 mM citrate, 100 mM NaCl, pH 5.0, resulting in a concentration of RNA cargo
Attorney Docket: 01155-0064-00PCT of approximately 0.45 mg/mL. The LNPs used contained ionizable lipid ((9Z,12Z)-3-((4,4- bis(octyloxy)butanoyl)oxy)-2-((((3-(diethylamino)propoxy)carbonyl)oxy)methyl)propyl octadeca-9,12-dienoate, also called 3-((4,4-bis(octyloxy)butanoyl)oxy)-2-((((3- (diethylamino)propoxy)carbonyl)oxy)methyl)propyl (9Z,12Z)-octadeca-9,12-dienoate), also called herein Lipid A, cholesterol, distearoylphosphatidylcholine (DSPC), and 1,2-dimyristoyl- rac-glycero-3-methoxypolyethylene glycol-2000 (PEG2K-DMG) (catalog # GM-020 from NOF, Tokyo, Japan) in a molar ratio as stated in each example. The LNPs were formulated with a lipid amine to RNA phosphate (N:P) molar ratio of about 6. The LNPs were prepared with a mixture of Cas9 mRNA and a guide RNA. [00466] The LNPs were prepared using a cross-flow technique utilizing impinging jet mixing of the lipid in ethanol with two volumes of RNA solution and one volume of water. First, the lipid in ethanol was mixed through a mixing cross with the two volumes of RNA solution. Then, a fourth stream of water was mixed with the outlet stream of the cross through an inline tee (See WO 2016/010840, FIG. 2). The LNPs were held for at least 1 hour at room temperature, and further diluted with water (approximately 1:1 v/v). Diluted LNPs were buffer exchanged into 50 mM Tris, 45 mM NaCl, 5% (w/v) sucrose, pH 7.5 (TSS) and concentrated as needed by methods known in the art. The resulting mixture was then filtered using a 0.2 μm sterile filter. The final LNPs were characterized to determine the encapsulation efficiency, polydispersity index, and average particle size. The final LNPs were stored at 4 °C or -80 °C until further use. 1.3 T Cell Preparation [00467] T cells were isolated from commercially obtained donor apheresis and cryopreserved. Upon thaw, T cells were plated at a density of 2.0 x 10^6 cells/mL in T cell growth media (TCGM) composed of CTS OpTmizer T Cell Expansion SFM and T Cell Expansion Supplement (ThermoFisher Cat. # A1048501) containing 2.5% human AB serum (GeminiBio, Cat. # 100-512), 1X Penicillin-Streptomycin (ThermoFisher, Cat. # 15140122), 1X Glutamax (ThermoFisher, Cat. # 35050061), and 10 mM HEPES (ThermoFisher, Cat. # 15630106) further supplemented with 200 U/mL recombinant human interleukin-2 (Peprotech, Cat. # 200-02), 5 ng/mL recombinant human interleukin-7 (Peprotech, Cat. # 200-07), and 5 ng/mL recombinant human interleukin-15 (Peprotech, Cat. # 200-15). T cells were activated with TransAct™ (1:100 dilution, Miltenyi Biotec, Cat. # 130-111-1).
Attorney Docket: 01155-0064-00PCT Example 2. T Cell Editing and Karyotyping [00468] Several features of T cell editing at the HLA-A locus were assessed for impact on protein knockdown via flow cytometry and on chromosomal integrity via karyotyping. Experiments examined the impact of HLA-A edit timing with respect to T cell activation and/or the impact of the lipid nanoparticle composition. 2.1 T cell editing [00469] T cells with the genotype indicated in Table 1 were isolated from human donor apheresis and were prepared and activated one day after thaw as described in Example 1. LNPs were formulated as described in Example 1 with the lipid molar ratio and RNA ratio indicated in this example. [00470] Table 1. T cell donor genotypes Donor HLA-A HLA-H

[ ] amp es were ac va e an e e on e schedule indicated in Table 2. Prior to editing, T cells were centrifuged, resuspended and plated at 1,000,000 cells/well in 1 mL/well in TCGM with 2.5% human AB serum on D0/D1 and resuspended at 500,000 cells/well in 1 mL/well in TCGM with 2.5% human AB serum on D3/D4. LNPs were prepared as described in Example 1 with a LNP composition A (LNP A) with a molar ratio of lipids of 50 Lipid A: 39.5 cholesterol: 9 DSPC: 1.5 PEG2k-DMG and with a ratio of gRNA to SpyCas9 mRNA of 1:2 by weight or LNP composition B (LNP B) with a molar ratio of lipids of 35 Lipid A: 47.5 cholesterol: 15 DSPC:2.5 PEG2k-DMG and with a ratio of gRNA to SpyCas9 mRNA of 1:1 by weight. LNPs were incubated with 2.5 ug/mL ApoE (Peprotech, Cat. # 350-02) in TCGM with 2.5% human AB serum at 37 °C for about 5 minutes. LNP A compositions were added to T cells at 5 μg total RNA cargo/ml each. LNP B compositions were added to T cells at 1 μg total RNA cargo/mL each. All edits for a given sample used LNPs with the same molar ratio of lipids throughout the experiment – all LNP A or all LNP B. Between edits, cells were incubated at 37 °C. HLA-A LNPs contained SpCas9 mRNA and gRNA G018995 (SEQ ID NO: 716) (targeting the genomic coordinates (hg38) chr6:29942864-29942884). CIITA LNPs contain SpCas9 mRNA and gRNA G013675 (SEQ ID NO: 724). TRAC LNPs contain SpCas9 mRNA and gRNA G013006 (SEQ ID NO: 708). “CAR AAV” in Table 2 indicates samples
Attorney Docket: 01155-0064-00PCT were treated with AAV6 virus at a multiplicity of infection (MOI) of 1 × 10^5. Samples labeled D0A, D0B, D1 #1 and D4 #1 were treated with 0.25 μM of Compound 4 in WO 2022/221696 at the same time as AAV administration. The AAV virus encoded a CD30-based chimeric antigen receptor (SEQ ID NO: 726) with homology arms flanking the gRNA G013006 cut site. [00472] Table 2. Editing schedule S
ample LNP C
omposition Day 0 Day 1 Day 3 Day 4 P
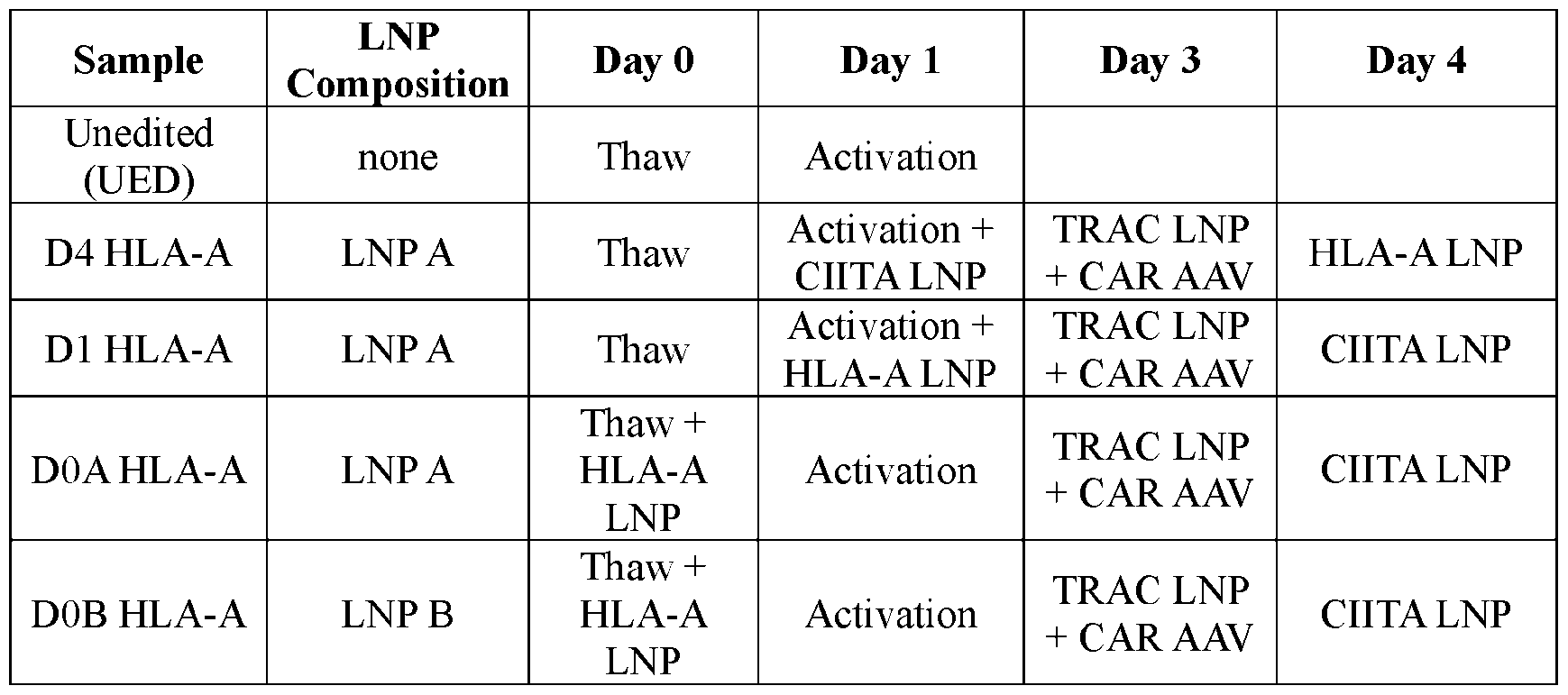
2.2 Flow cytometry [00473] T cells were analyzed by flow cytometry eleven days after thaw to determine surface biomarker expression for each target gene following editing. Cells were washed in FACS buffer (PBS + 2% FBS + 2 mM EDTA). T cells were incubated in 10 μg/mL CD30 Fc chimeric protein (R&D Systems, Cat. # 813-CD-100), washed, and then incubated in a cocktail of antibodies targeting HLA-A2 (Invitrogen, Cat. # 17-9876-42), HLA-A3 (BD Biosciences, Cat. # 747776), HLA-DR, DP, DQ (Biolegend, Cat. # 361712), CD3 (Biolegend, Cat. # 317336), CD4 (Biolegend, Cat. # 317434), CD8 (Biolegend, Cat. # 301046), and human IgG Fc (Biolegend, Cat. # 410708), and ViaKrome 808 Fixable Viability Dye (Beckman Coulter, Cat. # C36628). T cells were subsequently washed and analyzed on a Cytoflex instrument (Beckman Coulter). Data analysis was performed using FlowJo software package (v.10.6.1). T cells were gated on size, viability, CD4 or CD8 expression, and expression of markers indicated in Table 3. Flow cytometry data for CD8+ cells is shown in Table 3 and Fig. 1. CD3 expression is used as a biomarker for surface expression of the T cell receptor, indicating the cell is expressing protein from TRAC and TRBC1/2 loci. HLA-DP, DQ, DR expression is used as a biomarker for CIITA expression, as CIITA is a transcription factor that
Attorney Docket: 01155-0064-00PCT controls expression of these surface proteins. Comparable flow cytometry results were observed for CD8+ T cells and for CD4+ T cells. [00474] Table 3. The percentage of CD8+ cells positive for the indicated markers in Donor “1”or Donor “2” as measured by flow cytometry. H
LA-A3+ HLA-A2+ HLA-DQ, DP, D
R+ CD3+ IgG Fc+ .7 .1 .5 .4 .5 .8 .2
2.3 Karyotype Analysis [00475] Karyotype analysis was performed at KromaTiD, Inc. (Longmont, CO, USA) to assess chromosomal structure of chromosome 6, as HLA-A and HLA-H reside on chromosome 6 band p21.3. On day 11 or 12 post thaw, cells were harvested and cryopreserved for karyotyping and Droplet Digital PCR (ddPCR). KromaTiD prepared samples using standard G-banding techniques. Two hundred metaphase spreads per sample were analyzed for structural abnormalities in chromosome 6. Industry-standard protocols were used to score events (ISCN 2020: An International System for Human Cytogenomic Nomenclature, 2020). Table 4 shows the number of each type of chromosome 6 abnormality detected per sample. [00476] Table 4. Karyotype analysis indicating the number of spreads with the chromosome event indicated Donor Treatment Deletion (6)(p21.3)
Attorney Docket: 01155-0064-00PCT Donor Treatment Deletion (6)(p21.3) D0B HLA-A 1

. rop et gta C na ys s [00477] Droplet Digital PCR (ddPCR) was performed to determine copy number variation (CNV) of the short arm of human chromosome 6 (6p) following cell engineering. The NEDD9 gene, with a cytogenetic location of 6p24, was used as a marker for the distal portion of the short arm of human chromosome 6. The MRPL18 gene, with a cytogenetic location of 6q25.3, was used as a reference control on the long arm of human chromosome 6. [00478] Genomic DNA (gDNA) was isolated from cryopreserved engineered or untreated cells using the DNeasy Blood and Tissue Kits (Qiagen, Cat. # 69506) according to the manufacturer’s instructions and diluted using molecular grade distilled water (Gibco, Cat. # 46-00-CI). For the ddPCR assay, about 20 ng of gDNA from the engineered cells of Donor 2 was mixed with 2× ddPCR Supermix for Probes (Bio-Rad, Cat. # 1863024). Validated primers and probes for NEDD9 (Bio-Rad, Assay ID: dHsaCP1000467) and MRPL18 (Customized assay design, Bio-Rad) were added to final concentrations of 900 nM and 250 nM, respectively. Samples were processed with the Automated Droplet Generator (Bio-Rad, Cat. # 1864101). Droplet fluorescence was measured using QX200™ Droplet Reader (Bio-Rad, Cat. # 1864003). The measured copy number variation (CNV) for each treatment was obtained using the QuantaSoft™ Software, Regulatory Edition (Bio-Rad, Cat. # 1864011). The CNV was calculated using the formula: Copy Number Variation (CNV) = (NEDD9 copies/uL / MRPL18 copies/uL) * 2. Table 5 shows the CNV of the tested samples in triplicates. Cells with HLA-A editing on Day 4 of the engineering process showed a decrease copy number for the distal portion of the short arm of chromosome 6 compared to unedited cells or compared to cells with HLA-A editing on Day 0 or Day 1 of the engineering process. [00479] Table 5. ddPCR analysis showing the copy number variations of Chr6p in tested samples Donor Treatment CNV Mean SD

Attorney Docket: 01155-0064-00PCT Table 6. List of sequences [00480] In the following table and throughout, the terms “mA,” “mC,” “mU,” or “mG” are used to denote a nucleotide that has been modified with 2’-O-Me. [00481] In the following table, a “*” is used to depict a PS modification. In this application, the terms A*, C*, U*, or G* may be used to denote a nucleotide that is linked to the next (e.g., 3’) nucleotide with a PS bond. [00482] It is understood that if a DNA sequence (comprising Ts) is referenced with respect to an RNA, then Ts should be replaced with Us (which may be modified or unmodified depending on the context), and vice versa. [00483] In the following table, single amino acid letter code is used to provide peptide sequences. Description SEQ ID Sequence NO N U d 701707 m m C m m C m m C G C G G T C C G A C C T

Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO CTTCTGCCAGCAGTACCACACCTACCCCCTGACCTTCGGCGGCGGCA C A G G T C G A C G G S S N L R G G A C A A G T A G T G C G G G
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO CAAAGCTGCAGCTGAGCAAGGACACATACGACGACGACCTGGACAA G G G A A C T G G A T G C T G A A A G A A C A G A T A A G A A C A A T A A G G T T C
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO AGGGAAAGAGCAAGAAGCTGAAGAGCGTCAAGGAACTGCTGGGAA T C A C G T C A A A A G G T G C C G A C G C C G T A G C T A G T G A A A G T C T T G
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO ATCAACGGCATCCGGGACAAGCAGTCCGGCAAGACCATCCTGGACT C C T G G C T A A C A G T C G C G A G G G A G A C A G C C A C C G A A
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO CCACUUCCUGAUCGAGGGCGACCUGAACCCCGACAACUCCGACGUG C C C G C C C G A G C G G A C G C C G G A U C A A C G G C C C G C
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO UCAUGAACUUCUUCAAGACCGAGAUCACCCUGGCCAACGGCGAGA C A A G C G G C G G G C C A cc ga ac a ca cc a ga aa gg c g g c c c c c gc a c gc gc c g g ca a T c c ca T aa
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO ggTgaTgggccggcacaagcccgagaacaTcgTgaTcgagaTggcccgggagaaccagaccacccag cc c g T a ag ag c T g ag c g T c ag T T T g a ag a ag g gc g cc G C C C C A A C A G C C C T A C C C A A
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO ACCGCGAGAAGATCGAGAAGATCCTCACCTTCCGCATCCCCTACTAC G C T C A C C C G G G G G C G T C A C C C T C G C T G C G A C C G G C C A C C T T A G C
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO GCTACACCAGCACCAAGGAGGTCCTCGACGCCACCCTCATCCACCAG G G F E N E R F M E F R V K D Q L A A T D P R T V G F E N E R F M E F R V K D Q L A A T D P R
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO VILADANLDKVLSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDT V A A G A C C C T G A A A G T A G G T C G C G A G A A C A A G C G G C
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO ACGCCAGCCTGGGCACCTACCACGACCTGCTGAAGATCATCAAGGA C A A T C C C A G C A G G G G C A C C A A G T C A G G C G C C A C A G G C G C
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO GGACGACAGCTTCTTCCACCGGCTGGAGGAGAGCTTCCTGGTGGAGG A A G A T T A A G A T G A A C A A G G C C G C G G A G A G G T G G G A G A T
Attorney Docket: 01155-0064-00PCT Description SEQ ID Sequence NO CCTACCTGAACGCTGTGGTTGGCACGGCACTGATCAAGAAGTACCCC G C G C T C T G G C G G C A T A G G
Lipid Filing Compound Structure ID ID
Attorney Docket: 01155-0064-00PCT Lipid Filing Compound Structure ID ID Liid WO2020/ C d